4E3D
 
 | |
4E3E
 
 | | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl | | Descriptor: | MaoC domain protein dehydratase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl
To be Published
|
|
4E3F
 
 | |
4E3G
 
 | |
4E3H
 
 | |
4E3I
 
 | |
4E3J
 
 | |
4E3K
 
 | |
4E3L
 
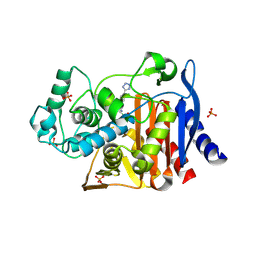 | |
4E3M
 
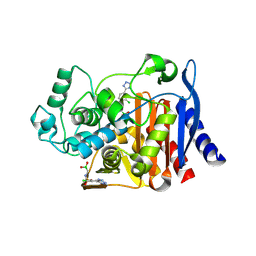 | |
4E3N
 
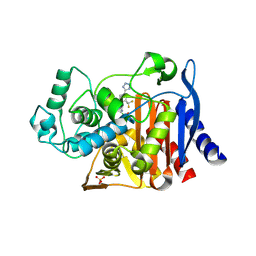 | |
4E3O
 
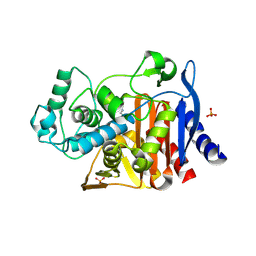 | |
4E3Q
 
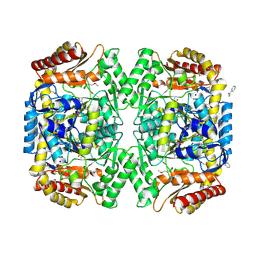 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3R
 
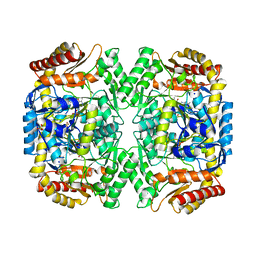 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3S
 
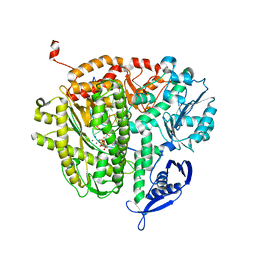 | | RB69 DNA Polymerase Ternary Complex with dQTP Opposite dT | | Descriptor: | 3-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-7-methyl-3H-imidazo[4,5-b]pyridine, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-03-10 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4E3T
 
 | |
4E3U
 
 | |
4E3V
 
 | |
4E3W
 
 | |
4E3X
 
 | | Crystal Structure of Mus musculus 1-pyrroline-5-carboxylate dehydrogenase cryoprotected in proline | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial, ... | | Authors: | Tanner, J.J, Pemberton, T.A. | | Deposit date: | 2012-03-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Proline: Mother Nature's cryoprotectant applied to protein crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E3Y
 
 | | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution
To be Published
|
|
4E3Z
 
 | | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | Putative oxidoreductase protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-11 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4E40
 
 | | The haptoglobin-hemoglobin receptor of Trypanosoma congolense | | Descriptor: | Putative uncharacterized protein | | Authors: | Higgins, M.K, Tkachenko, O, Brown, A, Reed, J, Carrington, M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the trypanosome haptoglobin-hemoglobin receptor and implications for nutrient uptake and innate immunity.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4E41
 
 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
4E42
 
 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | CHLORIDE ION, NITRATE ION, SODIUM ION, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
