1XC6
 
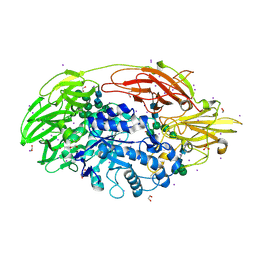 | | Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
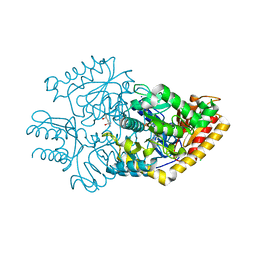
3R4T
 
 | |
3R57
 
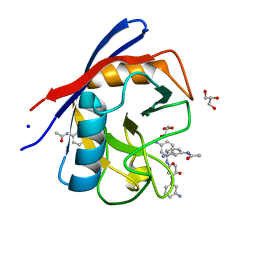 | | Human Cyclophilin D Complexed with a Fragment | | Descriptor: | GLYCEROL, N-(3-aminophenyl)acetamide, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
8H8Q
 
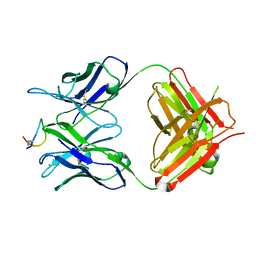 | | Fab-amyloid beta fragment complex at neutral pH | | Descriptor: | CHLORIDE ION, Fab, GLN-LYS-CYS-VAL-PHE-PHE-ALA-GLU-ASP-VAL-GLY-SER-ASN-CYS-GLY, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Matsushima, Y, Miki, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fab-amyloid beta fragment complex at neutral pH
To Be Published
|
|
3R0X
 
 | | Crystal structure of Selenomethionine incorporated apo D-serine deaminase from Salmonella tyhimurium | | Descriptor: | 1,2-ETHANEDIOL, D-serine dehydratase, SODIUM ION, ... | | Authors: | Bharath, S.R, Shveta, B, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Febs J., 2011
|
|
8FHJ
 
 | | Crystal structure of a FAD monooxygenease from Methylocystis sp. Strain SB2 | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Stewart, A.M, Sawaya, M.R, Stewart, C.E. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of MbnF: an NADPH-dependent flavin monooxygenase from Methylocystis strain SB2.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3RC9
 
 | |
1K4D
 
 | | Potassium Channel KcsA-Fab complex in low concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
8G2E
 
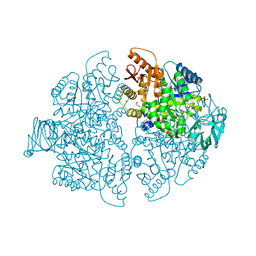 | | PKM2 bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-aminophenyl)methyl]-5-methyl-7-[methyl(oxidanyl)-$l^{3}-sulfanyl]pyridazino[4,5-b]indol-4-one, MAGNESIUM ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Development of Novel Small-Molecule Activators of Pyruvate Kinase Muscle Isozyme 2, PKM2, to Reduce Photoreceptor Apoptosis.
Pharmaceuticals, 16, 2023
|
|
8HCG
 
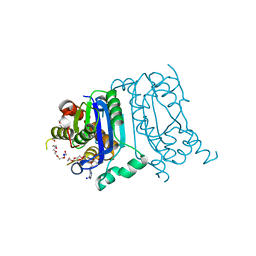 | | Crystal structure of mTREX1-dAMP complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCC
 
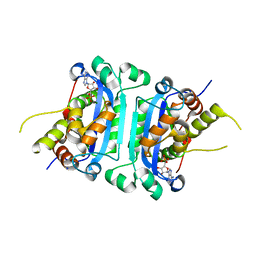 | | Crystal structure of mTREX1-RNA product complex (AMP) | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
3R2O
 
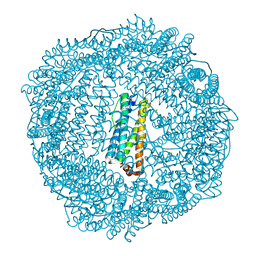 | | 1.95 A resolution structure of As-Isolated FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, SODIUM ION, SULFATE ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8HV1
 
 | | Crystal structure of EGFR_DMX in complex with covalently bound fragment 1 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-(3-bromophenyl)-1,4-dihydro-1,2,4-triazole-5-thione, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
4HZY
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
1XJ4
 
 | | CO-bound structure of BjFixLH | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Key, J, Moffat, K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Deoxy and CO-Bound bjFixLH Reveal Details of Ligand Recognition and Signaling
Biochemistry, 44, 2005
|
|
1JG8
 
 | | Crystal Structure of Threonine Aldolase (Low-specificity) | | Descriptor: | CALCIUM ION, L-allo-threonine aldolase, SODIUM ION | | Authors: | Kielkopf, C.L, Bonanno, J, Ray, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-23 | | Release date: | 2001-07-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Low-specificity Threonine Aldolase, a Key Enzyme in Glycine Biosynthesis
To be published
|
|
8HU4
 
 | | Limosilactobacillus reuteri N1 GtfB | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-22 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8HUF
 
 | | B28 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-chlorophenyl)carbonylamino]-4-[4-(2,5-dimethylphenyl)piperazin-1-yl]benzoic acid, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
1JN9
 
 | |
3SHX
 
 | | Frog M-ferritin with magnesium, L134P mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Tosha, T, Ng, H.L, Alber, T, Theil, E.C. | | Deposit date: | 2011-06-17 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Frog M-ferritin with magnesium, L134P mutant
To be Published
|
|
8HW3
 
 | | Limosilactobacillus reuteri N1 GtfB-acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
4HZX
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
1JRX
 
 | | Crystal structure of Arg402Ala mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
