1GCZ
 
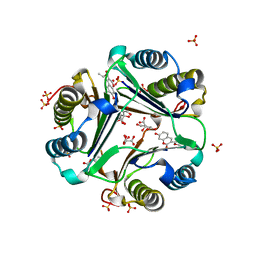 | | MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) COMPLEXED WITH INHIBITOR. | | Descriptor: | 7-HYDROXY-2-OXO-CHROMENE-3-CARBOXYLIC ACID ETHYL ESTER, CITRIC ACID, MACROPHAGE MIGRATION INHIBITORY FACTOR, ... | | Authors: | Katayama, N, Kurihara, H. | | Deposit date: | 2000-08-24 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography.
J.Med.Chem., 44, 2001
|
|
1G8C
 
 | |
1GD0
 
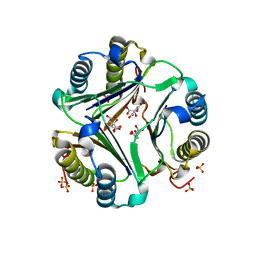 | | HUMAN MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) | | Descriptor: | CITRIC ACID, MACROPHAGE MIGRATION INHIBITORY FACTOR, SULFATE ION | | Authors: | Kurihara, H, Katayama, N. | | Deposit date: | 2000-08-24 | | Release date: | 2001-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography.
J.Med.Chem., 44, 2001
|
|
1JB2
 
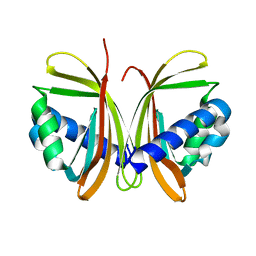 | | CRYSTAL STRUCTURE OF NTF2 M84E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1E5C
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
3RMJ
 
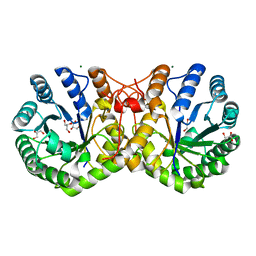 | | Crystal structure of truncated alpha-Isopropylmalate Synthase from Neisseria meningitidis | | Descriptor: | 2-isopropylmalate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Huisman, F.H.A, Baker, H.M, Koon, N, Baker, E.N, Parker, E.J. | | Deposit date: | 2011-04-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Removal of the C-terminal regulatory domain of alpha-isopropylmalate synthase disrupts functional substrate binding
Biochemistry, 51, 2012
|
|
1JEZ
 
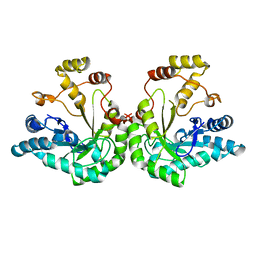 | | THE STRUCTURE OF XYLOSE REDUCTASE, A DIMERIC ALDO-KETO REDUCTASE FROM CANDIDA TENUIS | | Descriptor: | XYLOSE REDUCTASE | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-06-19 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
3R98
 
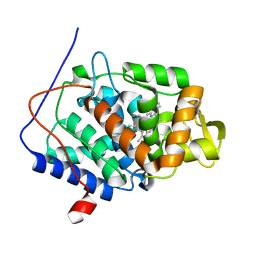 | | Joint Neutron and X-ray structure of Cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blakeley, M.P, Fisher, S.J, Gumiero, A, Moody, P.C.E, Raven, E.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2.4 Å), X-RAY DIFFRACTION | | Cite: | Hydrogen bonds in heme peroxidases: a combined X-ray and neutron study of cytochrome c peroxidase
To be Published
|
|
3RQJ
 
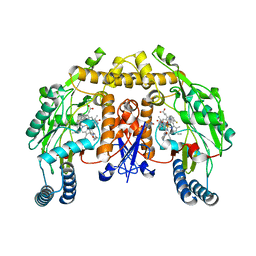 | | Structure of the neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((1S,2R)-2-(3-Fluorophenyl)cyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(1S,2R)-2-(3-fluorophenyl)cyclopropyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2011-04-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cyclopropyl- and methyl-containing inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 21, 2013
|
|
3PL7
 
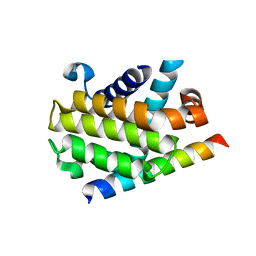 | |
1HTD
 
 | | STRUCTURAL INTERACTION OF NATURAL AND SYNTHETIC INHIBITORS WITH THE VENOM METALLOPROTEINASE, ATROLYSIN C (HT-D) | | Descriptor: | ATROLYSIN C, CALCIUM ION, ZINC ION | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1994-01-20 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3PMH
 
 | |
3PNH
 
 | | Structure of Bovine Endothelial Nitric Oxide Synthase Heme Domain in complex with 6-(((3R,4R)-4-(2-((2-FLUORO-2-(3-FLUOROPHENYL) ETHYL)AMINO)ETHOXY)PYRROLIDIN-3-YL)METHYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-2-fluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
3QJX
 
 | | Crystal Structure of E. coli Aminopeptidase N in complex with L-Serine | | Descriptor: | Aminopeptidase N, GLYCEROL, MALONATE ION, ... | | Authors: | Addlagatta, A, Gumpena, R, Kishor, C, Ganji, R.J. | | Deposit date: | 2011-01-31 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of alpha, beta- and alpha, gamma-Diamino Acid Scaffolds for the Inhibition of M1 Family Aminopeptidases
Chemmedchem, 6, 2011
|
|
3Q9A
 
 | | Structure of neuronal nitric oxide synthase in the ferric state in complex with N-5-[2-(ethylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Doukov, T, Poulos, T.L. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Temperature-dependent spin crossover in neuronal nitric oxide synthase bound with the heme-coordinating thioether inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
1G89
 
 | |
3QJT
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
1J4V
 
 | | CYANOVIRIN-N | | Descriptor: | CYANOVIRIN-N | | Authors: | Clore, G.M, Bewley, C.A. | | Deposit date: | 2001-11-21 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings.
J.Magn.Reson., 154, 2002
|
|
1FW5
 
 | | SOLUTION STRUCTURE OF MEMBRANE BINDING PEPTIDE OF SEMLIKI FOREST VIRUS MRNA CAPPING ENZYME NSP1 | | Descriptor: | NONSTRUCTURAL PROTEIN NSP1 | | Authors: | Lampio, A, Kilpelinen, I, Pesonen, S, Karhi, K, Auvinen, P, Somerharju, P, Kriinen, L. | | Deposit date: | 2000-09-21 | | Release date: | 2001-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane binding mechanism of an RNA virus-capping enzyme.
J.Biol.Chem., 275, 2001
|
|
3Q99
 
 | | Structure of neuronal nitric oxide synthase in the ferric state in complex with N~5~-[(3-(ethylsulfanyl)propanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Doukov, T, Poulos, T.L. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Temperature-dependent spin crossover in neuronal nitric oxide synthase bound with the heme-coordinating thioether inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
3RQO
 
 | | Structure of the endothelial nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((1S,2R/1R,2S)-2-(3-Clorophenyl)cyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(1R,2S)-2-(3-chlorophenyl)cyclopropyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2011-04-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Cyclopropyl- and methyl-containing inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 21, 2013
|
|
1E8L
 
 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
1KLL
 
 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1ERJ
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL WD40 DOMAIN OF TUP1 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR TUP1 | | Authors: | Sprague, E.R, Redd, M.J, Johnson, A.D, Wolberger, C. | | Deposit date: | 2000-04-06 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast.
EMBO J., 19, 2000
|
|
1G3P
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAINS OF BACTERIOPHAGE MINOR COAT PROTEIN G3P | | Descriptor: | MINOR COAT PROTEIN, SULFATE ION | | Authors: | Lubkowski, J, Hennecke, F, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1997-12-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p.
Nat.Struct.Biol., 5, 1998
|
|
