2BKJ
 
 | |
5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
1BL3
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BL5
 
 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | Descriptor: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1998-07-23 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
3CRD
 
 | | NMR STRUCTURE OF THE RAIDD CARD DOMAIN, 15 STRUCTURES | | Descriptor: | RAIDD | | Authors: | Chou, J.J, Matsuo, H, Duan, H, Wagner, G. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | Descriptor: | LQH III ALPHA-LIKE TOXIN | | Authors: | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
1LU1
 
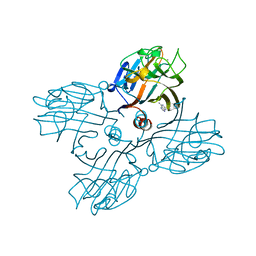 | | THE STRUCTURE OF THE DOLICHOS BIFLORUS SEED LECTIN IN COMPLEX WITH THE FORSSMAN DISACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ADENINE, CALCIUM ION, ... | | Authors: | Hamelryck, T.W, Loris, R, Bouckaert, J, Strecker, G, Imberty, A, Fernandez, E, Wyns, L, Etzler, M.E. | | Deposit date: | 1998-07-24 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1EER
 
 | |
4ALD
 
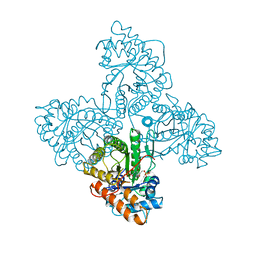 | | HUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATE | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-BISPHOSPHATE ALDOLASE | | Authors: | Dalby, A.R, Dauter, Z, Littlechild, J.A. | | Deposit date: | 1998-07-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications.
Protein Sci., 8, 1999
|
|
2EZZ
 
 | |
2EZX
 
 | |
2EZY
 
 | |
1BM2
 
 | |
1BMZ
 
 | |
1BMX
 
 | |
1BMW
 
 | | A fibronectin type III fold in plant allergens: The solution structure of Phl PII from timothy grass pollen, NMR, 38 STRUCTURES | | Descriptor: | POLLEN ALLERGEN PHL P2 | | Authors: | De Marino, S, Morelli, M.A.C, Fraternali, F, Tamborino, E, Vrtala, S, Dolecek, C, Arosio, P, Valenta, R, Pastore, A. | | Deposit date: | 1998-07-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An Immunoglobulin-Like Fold in a Major Plant Allergen: The Solution Structure of Phl P 2 from Timothy Grass Pollen.
Structure Fold.Des., 7, 1999
|
|
1CPX
 
 | |
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BM5
 
 | |
1BM4
 
 | | MOMLV CAPSID PROTEIN MAJOR HOMOLOGY REGION PEPTIDE ANALOG | | Descriptor: | PROTEIN (MOLONEY MURINE LEUKEMIA VIRUS CAPSID) | | Authors: | Clish, C.B, Peyton, D.H, Barklis, E. | | Deposit date: | 1998-07-28 | | Release date: | 1998-08-05 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human immunodeficiency virus type 1 (HIV-1) and moloney murine leukemia virus (MoMLV) capsid protein major-homology-region peptide analogs by NMR spectroscopy.
Eur.J.Biochem., 257, 1998
|
|
1BRR
 
 | | X-RAY STRUCTURE OF THE BACTERIORHODOPSIN TRIMER/LIPID COMPLEX | | Descriptor: | 3,7,11,15-TETRAMETHYL-HEXADECAN-1-OL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Essen, L.-O, Siegert, R, Oesterhelt, D. | | Deposit date: | 1998-07-28 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3PYP
 
 | | PHOTOACTIVE YELLOW PROTEIN, CRYOTRAPPED EARLY LIGHT CYCLE INTERMEDIATE | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Soltis, S.M, Kuhn, P, Canestrelli, I.L, Getzoff, E.D. | | Deposit date: | 1998-07-28 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure at 0.85 A resolution of an early protein photocycle intermediate.
Nature, 392, 1998
|
|
3KTQ
 
 | |
3LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS WITH PHOSPHOCHOLINE BOUND | | Descriptor: | LEUKOCIDIN F SUBUNIT, PHOSPHOCHOLINE | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
