1X5Y
 
 | | Solution structure of the fibronectin type-III domain of mouse myosin-binding protein C, Fast-type homolog | | Descriptor: | myosin binding protein C, fast-type | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibronectin type-III domain of mouse myosin-binding protein C, Fast-type homolog
To be Published
|
|
3II3
 
 | | Structure of ORF157 from Acidianus filamentous Virus 1 | | Descriptor: | GLYCEROL, NICKEL (II) ION, Putative uncharacterized protein | | Authors: | Goulet, A, Redder, P, Pina, M, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-07-31 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
6FYP
 
 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/CX-4945 AT 2.29A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
7M09
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with blunt-ended DSB substrate and incoming dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0B
 
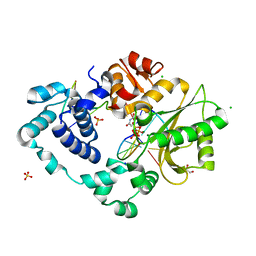 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound mismatched DSB and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7DCR
 
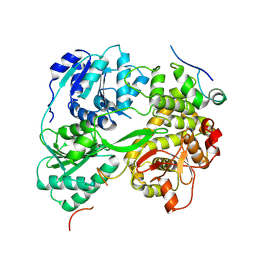 | | cryo-EM structure of the DEAH-box helicase Prp2 in complex with its coactivator Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2 | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
8S84
 
 | |
3IJ2
 
 | | Ligand-receptor structure | | Descriptor: | Beta-nerve growth factor, Nerve growth factor receptor (TNFR superfamily, member 16) | | Authors: | Feng, D, Garcia, K.C. | | Deposit date: | 2009-08-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Molecular and structural insight into proNGF engagement of p75NTR and sortilin.
J.Mol.Biol., 396, 2010
|
|
8SRU
 
 | |
2UXY
 
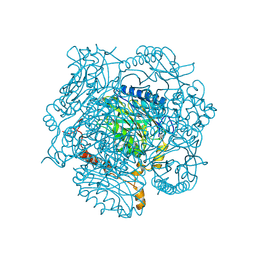 | | Aliphatic amidase | | Descriptor: | ALIPHATIC AMIDASE, SULFATE ION | | Authors: | Andrade, J, KArmali, A, Carrondo, M.A, Frazao, C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Amidase from Pseudomonas Aeruginosa Showing a Trapped Acyl Transfer Reaction Intermediate State.
J.Biol.Chem., 282, 2007
|
|
7M07
 
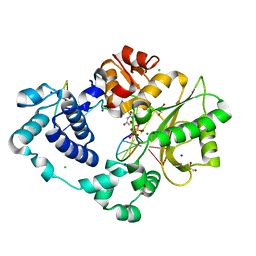 | | Pre-catalytic ternary complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
8SRW
 
 | |
8SRV
 
 | |
1WO9
 
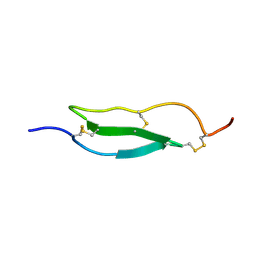 | | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop | | Descriptor: | trypsin inhibitor | | Authors: | Kellenberger, C, Ferrat, G, Leone, P, Darbon, H, Roussel, A. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop
Biochemistry, 42, 2003
|
|
8OF0
 
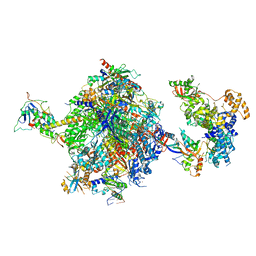 | | Structure of the mammalian Pol II-SPT6-Elongin complex, Structure 1 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEV
 
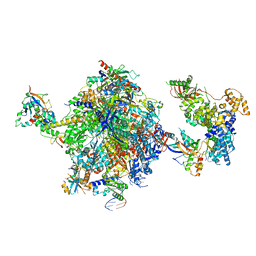 | | Structure of the mammalian Pol II-SPT6-Elongin complex, lacking ELOA latch (composite structure, structure 3) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEW
 
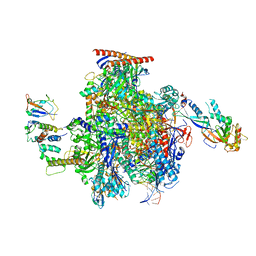 | | Structure of the mammalian Pol II-Elongin complex, lacking the ELOA latch (composite structure, structure 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4R7E
 
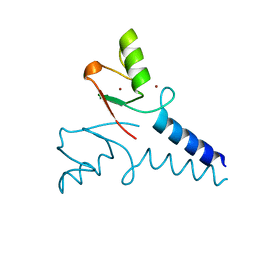 | | Structure of Bre1 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1, ZINC ION | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of the yeast Bre1 RING domain.
Proteins, 83, 2015
|
|
2XQ0
 
 | | Structure of yeast LTA4 hydrolase in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CHLORIDE ION, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
5WKN
 
 | | Crystal structure of the parainfluenza virus 5 nucleoprotein-phosphoprotein complex | | Descriptor: | Nucleoprotein, Phosphoprotein | | Authors: | Aggarwal, M, Leser, G.P, Kors, C.A, Lamb, R.A. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the Paramyxovirus Parainfluenza Virus 5 Nucleoprotein in Complex with an Amino-Terminal Peptide of the Phosphoprotein.
J. Virol., 92, 2018
|
|
4Y5I
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 126B with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Microtubule-associated protein tau | | Authors: | Leysen, S, Bartel, M, Milroy, L, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
8XUV
 
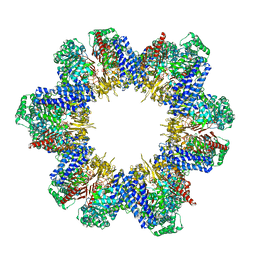 | | Cryo-EM structure of tomato NRC2 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
2VO4
 
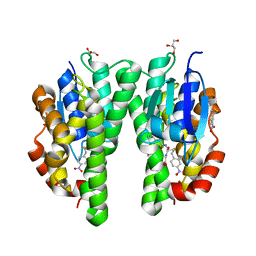 | | Glutathione transferase from Glycine max | | Descriptor: | 2,4-D INDUCIBLE GLUTATHIONE S-TRANSFERASE, 4-NITROPHENYL METHANETHIOL, GLYCEROL, ... | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and Functional Characterization of the Fluorodifen-Inducible Glutathione Transferase from Glycine Max Reveals an Active Site Topography Suited for Diphenylether Herbicides and a Novel L-Site.
J.Mol.Biol., 385, 2009
|
|
7B1H
 
 | |
