1ONM
 
 | | Solution Structure of a DNA duplex containing A:G mismatch. d(GCTTCAGTCGT):d(ACGACGGAAGC) | | Descriptor: | 5'-D(*AP*CP*GP*AP*CP*GP*GP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*CP*AP*GP*TP*CP*GP*T)-3' | | Authors: | Sanchez, A.M, Volk, D.E, Gorenstein, D.G, Lloyd, R.S. | | Deposit date: | 2003-02-28 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Initiation of repair of A/G mismatches is modulated by sequence context
DNA REPAIR, 2, 2003
|
|
5S18
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3QUX
 
 | | Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
190D
 
 | |
4CBZ
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN JAGGED-1, alpha-L-fucopyranose | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
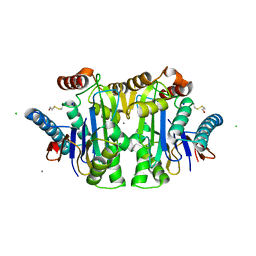
1JN9
 
 | |
3B4C
 
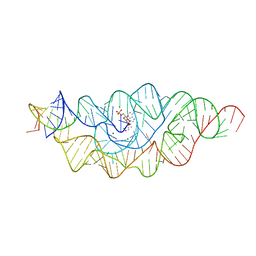 | |
1AA5
 
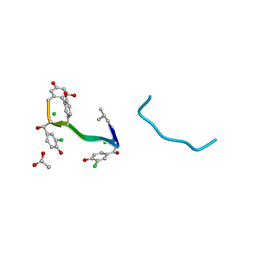 | | VANCOMYCIN | | Descriptor: | ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Bevivino, A.E, Korty, B.D, Axelsen, P.H. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Simultaneous Recognition of a Carboxylate-Containing Ligand and an Intramolecular Surrogate Ligand in the Crystal Structure of an Asymmetric Vancomycin Dimer.
J.Am.Chem.Soc., 119, 1997
|
|
1AFE
 
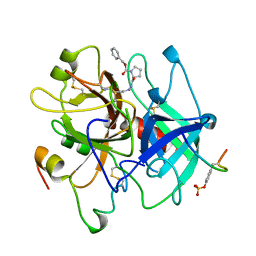 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
1TOB
 
 | | SACCHARIDE-RNA RECOGNITION IN AN AMINOGLYCOSIDE ANTIBIOTIC-RNA APTAMER COMPLEX, NMR, 7 STRUCTURES | | Descriptor: | 1,3-DIAMINO-5,6-DIHYDROXYCYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Suri, A.K, Fiala, R, Patel, D.J. | | Deposit date: | 1996-12-12 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex.
Chem.Biol., 4, 1997
|
|
4J7A
 
 | | Crystal Structure of Est25 - a Bacterial Homolog of Hormone-Sensitive Lipase from a Metagenomic Library | | Descriptor: | Esterase | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Park, K.S, Joo, S.B, Kim, K.K, Kim, D.H. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library
Acta Crystallogr.,Sect.D, 69, 2014
|
|
1AHP
 
 | | OLIGOSACCHARIDE SUBSTRATE BINDING IN ESCHERICHIA COLI MALTODEXTRIN PHSPHORYLASE | | Descriptor: | E.COLI MALTODEXTRIN PHOSPHORYLASE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | O'Reilly, M, Watson, K.A, Schinzel, R, Palm, D, Johnson, L.N. | | Deposit date: | 1997-04-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oligosaccharide substrate binding in Escherichia coli maltodextrin phosphorylase.
Nat.Struct.Biol., 4, 1997
|
|
4AFK
 
 | | In meso structure of alginate transporter, AlgE, from Pseudomonas aeruginosa, PAO1 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Tan, J, Pye, V.E, Aragao, D, Caffrey, M. | | Deposit date: | 2012-01-19 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A Conformational Landscape for Alginate Secretion Across the Outer Membrane of Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BAJ
 
 | | Human Pancreatic Alpha-Amylase in Complex with Nitrate and Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
1K26
 
 | | Structure of a Nudix Protein from Pyrobaculum aerophilum Solved by the Single Wavelength Anomolous Scattering Method | | Descriptor: | ACETIC ACID, GLYCEROL, IRIDIUM (III) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1A6A
 
 | | THE STRUCTURE OF AN INTERMEDIATE IN CLASS II MHC MATURATION: CLIP BOUND TO HLA-DR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ghosh, P, Amaya, M, Mellins, E, Wiley, D.C. | | Deposit date: | 1998-02-22 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3.
Nature, 378, 1995
|
|
3RKE
 
 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
1FCX
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARGAMMA-SELECTIVE RETINOID BMS184394 | | Descriptor: | 6-[HYDROXY-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALEN-2-YL)-METHYL]-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1LVH
 
 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
1MCC
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
5VYS
 
 | | Crystal structure of the WbkC N-formyltransferase (C47S variant) from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
3OAU
 
 | | Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Huber, M, Wilson, I.A, Burton, D.R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged.
J.Virol., 84, 2010
|
|
3BAI
 
 | | Human Pancreatic Alpha Amylase with Bound Nitrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRATE ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
1M6K
 
 | | Structure of the OXA-1 class D beta-lactamase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, beta-lactamase OXA-1 | | Authors: | Sun, T, Nukaga, M, Mayama, K, Braswell, E.H, Knox, J.R. | | Deposit date: | 2002-07-16 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of beta-lactamases of classes A and D: 1.5A crystallographic structure of the class D OXA-1 oxacillinase
PROTEIN SCI., 12, 2003
|
|
