4A84
 
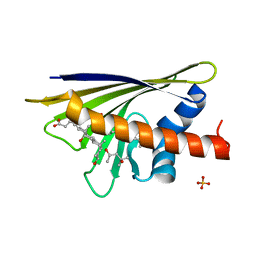 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a F30V mutant in complex with deoxycholate. | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
1O8P
 
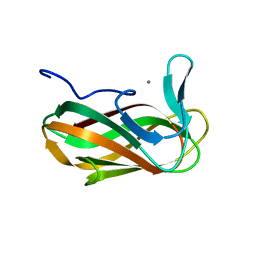 | | Unbound structure of CsCBM6-3 from Clostridium stercorarium | | Descriptor: | CALCIUM ION, PUTATUVE ENDO-XYLANASE | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains.
J. Mol. Biol., 327, 2003
|
|
5NMR
 
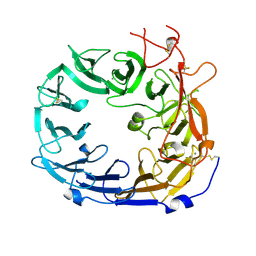 | | Monomeric mouse Sortilin extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Leloup, N.O.L, Loessl, P, Meijer, D.H.M, Heck, A.J.R, Thies-Weesie, D.M.E, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
4A87
 
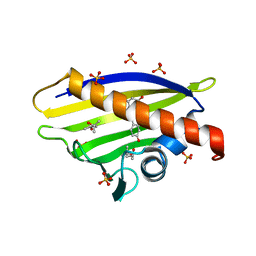 | |
4A81
 
 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in ternary complex with 8-Anilinonaphthalene-1-sulfonate (ANS) and deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
4PP0
 
 | | Structure of the PBP NocT-M117N in complex with pyronopaline | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1S)-4-carbamimidamido-1-carboxybutyl]-5-oxo-D-proline, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
4A88
 
 | |
4A83
 
 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with deoxycholate. | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
1XKI
 
 | | Crystal structure of human tear lipocalin/von Ebners gland protein | | Descriptor: | CHLORIDE ION, Von Ebner's gland protein, ZINC ION | | Authors: | Breustedt, D.A, Korndoerfer, I.P, Redl, B, Skerra, A. | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8-A crystal structure of human tear lipocalin reveals an extended branched cavity with capacity for multiple ligands
J.Biol.Chem., 280, 2005
|
|
5IF6
 
 | |
3I7L
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of DDB2 | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4A8G
 
 | |
4WQP
 
 | |
4A80
 
 | |
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
3LSD
 
 | | N-Domain of human adhesion/growth-regulatory galectin-9 | | Descriptor: | Galectin-9 | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2010-02-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution.
Int.J.Biochem.Cell Biol., 42, 2010
|
|
4A85
 
 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with kinetin. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
3HT8
 
 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUA
 
 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3ZM5
 
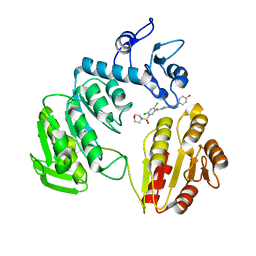 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | 2,4-bis(chloranyl)-N-[3-cyano-6-[(4-hydroxyphenyl)methyl]-5,7-dihydro-4H-thieno[2,3-c]pyridin-2-yl]-5-morpholin-4-ylsulfonyl-benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
3HU8
 
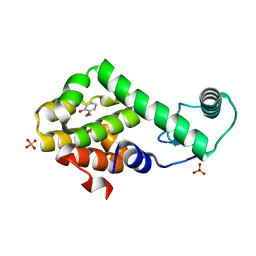 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
7L4Z
 
 | | Structure of SARS-CoV-2 spike RBD in complex with cyclic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2, Spike protein S1 | | Authors: | Christie, M, Mackay, J.P, Passioura, T, Payne, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Discovery of Cyclic Peptide Ligands to the SARS-CoV-2 Spike Protein Using mRNA Display.
Acs Cent.Sci., 7, 2021
|
|
3I7K
 
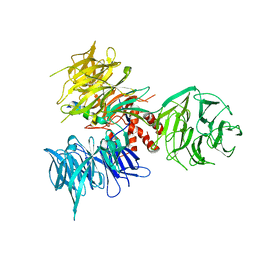 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WHX | | Descriptor: | DNA damage-binding protein 1, X protein | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I8E
 
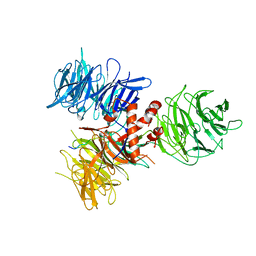 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR42A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 42A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
