4XJB
 
 | | X-ray structure of Lysozyme1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJG
 
 | | X-ray structure of Lysozyme B2 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJI
 
 | | X-ray structure of LysozymeS2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7CLT
 
 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
7LOU
 
 | | Crystal structure of Clostridium difficile Toxin B (TcdB) glucosyltransferase in complex with UDP and isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Glucosyltransferase TcdB, ... | | Authors: | Harijan, R.K, Paparella, A.S, Aboulache, B.L, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Inhibition of Clostridium difficile TcdA and TcdB toxins with transition state analogues.
Nat Commun, 12, 2021
|
|
5UIN
 
 | | X-ray structure of the W305A variant of the FdtF N-formyltransferase from salmonella enteric O60 | | Descriptor: | CHLORIDE ION, Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
6Z7K
 
 | | Crystal structure of CTX-M-15 in complex with the imine form of hydrolysed tazobactam | | Descriptor: | (2~{S},3~{S})-3-[bis(oxidanylidene)-$l^{5}-sulfanyl]-3-methyl-2-[(~{E})-3-oxidanylidenepropylideneamino]-4-(1,2,3-triaz ol-1-yl)butanoic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 2022
|
|
2WLC
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
5UIL
 
 | | X-ray structure of the FdtF N-formyltransferase from Salmonella enterica O60 in complex with TDP-Fuc3N and tetrahydrofolate | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, Formyltransferase, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
1URR
 
 | |
4XJD
 
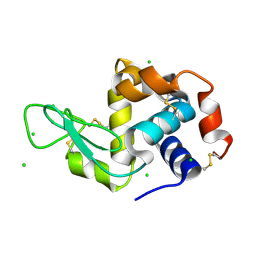 | | X-ray structure of Lysozyme2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJH
 
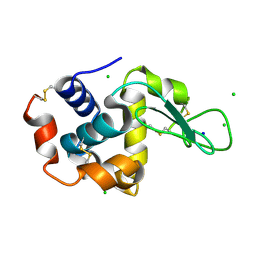 | | X-ray structure of LysozymeS1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1V0N
 
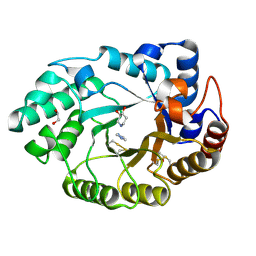 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
4XJN
 
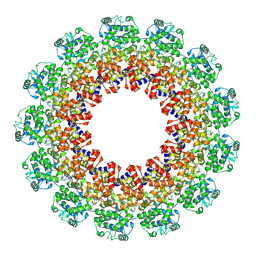 | | Structure of the parainfluenza virus 5 nucleocapsid-RNA complex: an insight into paramyxovirus polymerase activity | | Descriptor: | LEAD (II) ION, Nucleocapsid, RNA (78-MER) | | Authors: | Alayyoubi, M, Leser, G.P, Kors, C.A, Lamb, R.A. | | Deposit date: | 2015-01-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6U2O
 
 | |
3I6S
 
 | | Crystal Structure of the plant subtilisin-like protease SBT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Subtilisin-like protease, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rose, R, Ottmann, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7V3D
 
 | |
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
7VMI
 
 | |
6F3X
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
4X9Z
 
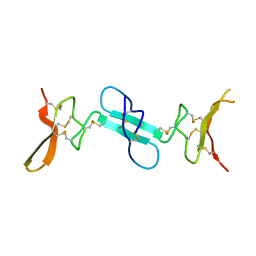 | | Dimeric conotoxin alphaD-GeXXA | | Descriptor: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | Authors: | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
1V0L
 
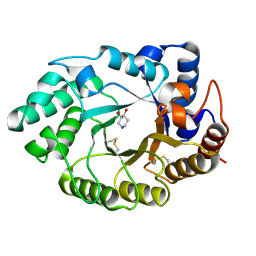 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
7CPO
 
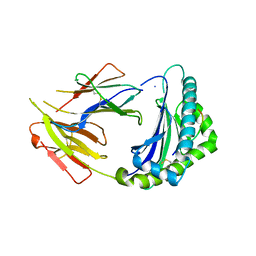 | | Crystal Structure of Anolis carolinensis MHC I complex | | Descriptor: | HIS-VAL-TYR-GLY-PRO-LEU-LYS-PRO-ILE, MHC CLASS I ANTIGEN, beta2-microglobulin | | Authors: | Wang, Y, Qu, Z, Ma, L, Wei, X, Zhang, N, Xia, C. | | Deposit date: | 2020-08-07 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the MHC Class I (MHC-I) Molecule in the Green Anole Lizard Demonstrates the Unique MHC-I System in Reptiles.
J Immunol., 206, 2021
|
|
1V0H
 
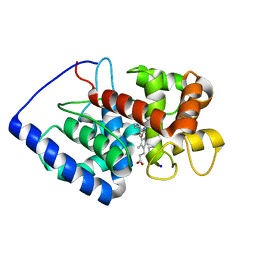 | | ASCOBATE PEROXIDASE FROM SOYBEAN CYTOSOL IN COMPLEX WITH SALICYLHYDROXAMIC ACID | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SALICYLHYDROXAMIC ACID, ... | | Authors: | Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Ascorbate Peroxidase-Salicylhydroxamic Acid Complex
Biochemistry, 43, 2004
|
|
3HYJ
 
 | | Crystal structure of the N-terminal LAGLIDADG domain of DUF199/WhiA | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DUF199/WhiA, ... | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease.
Structure, 17, 2009
|
|
