7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ8
 
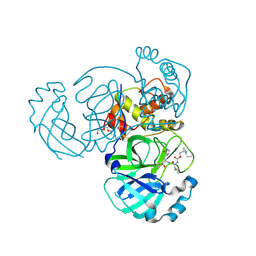 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 14d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2S)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, Orf1a protein, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ2
 
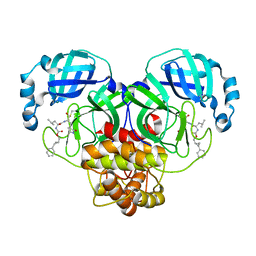 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ3
 
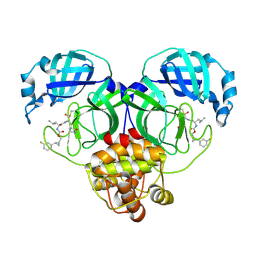 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7WQV
 
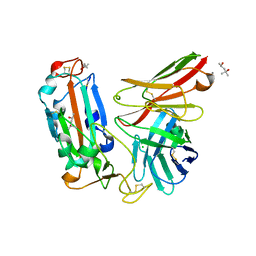 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
7TQ4
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TPR
 
 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
7WPV
 
 | | Fab14 - a SARS-CoV2 RBD neutralising antibody | | Descriptor: | Fab14 heavy chain, Fab14 light chain | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7TP3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WPH
 
 | | SARS-CoV2 RBD bound to Fab06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7WOP
 
 | | The local refined map of Omicron spike with bispecific antibody FD01 | | Descriptor: | 16L9, GW01, Spike protein S1 | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOR
 
 | | The state 2 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOS
 
 | | The state 3 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOV
 
 | | The state 5 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOU
 
 | | The state 4 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOW
 
 | | The state 6 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, GW01 Fv, Spike glycoprotein | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.11 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOQ
 
 | | The state 1 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOB
 
 | | SARS-CoV-2 Spike in complex with IgG 553-60 (2-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOG
 
 | | SARS-CoV-2 Omicron S monomer complexed with 553-49 | | Descriptor: | 553-49 VH, 553-49 VL, Spike protein S1 | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOC
 
 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOA
 
 | | SARS-CoV-2 Spike in complex with IgG 553-60 (1-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
