7PKP
 
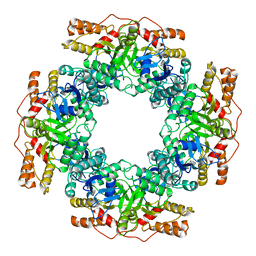 | | NSP2 RNP complex | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PT0
 
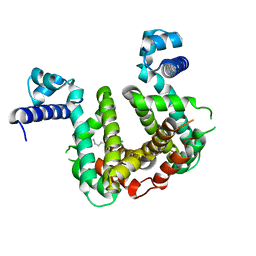 | | SCO3201 with putative ligand | | Descriptor: | SPERMIDINE, TetR family transcriptional regulator | | Authors: | Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2021-09-25 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 290, 2023
|
|
7ZTL
 
 | |
8A6I
 
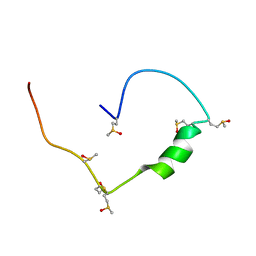 | | Structure of the low complexity domain of TDP-43 (fragment 309-350) with methionine sulfoxide modifications | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Carrasco, J, Anton, R, Pantoja-Uceda, D, Laurents, D.V, Oroz, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-08 | | Method: | SOLUTION NMR | | Cite: | Metamorphism in TDP-43 prion-like domain determines chaperone recognition.
Nat Commun, 14, 2023
|
|
7ZGJ
 
 | |
7ZGK
 
 | |
1OFE
 
 | |
7ZRB
 
 | |
1OMQ
 
 | |
7ZQT
 
 | |
5T8A
 
 | | Recombinant cytotoxin-I from the venom of cobra N. oxiana | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
5TDQ
 
 | | Crystal Structure of the GOLD domain of ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | McPhail, J.A, Burke, J.E. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | The Molecular Basis of Aichi Virus 3A Protein Activation of Phosphatidylinositol 4 Kinase III beta , PI4KB, through ACBD3.
Structure, 25, 2017
|
|
5TEZ
 
 | | TCR F50 recgonizing M1-HLA-A2 | | Descriptor: | Beta-2-microglobulin, GLY-ILE-LEU-GLY-PHE-VAL-PHE-THR-LEU, HLA class I histocompatibility antigen, ... | | Authors: | Yang, X, Mariuzza, R.A. | | Deposit date: | 2016-09-23 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for clonal diversity of the human T-cell response to a dominant influenza virus epitope.
J. Biol. Chem., 292, 2017
|
|
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
8AG3
 
 | |
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
5UFV
 
 | |
5UGQ
 
 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
8AXH
 
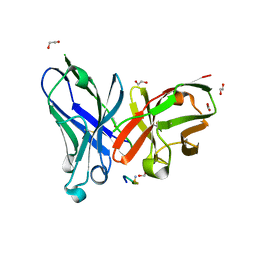 | | Crystal structure of a MUC1-like glycopeptide containing the unnatural L-4-hydroxynorvaline in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Mucin-1 subunit alpha, ... | | Authors: | Bermejo, I, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2022-08-31 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Approach for the Development of MUC1-Glycopeptide-Based Cancer Vaccines with Predictable Responses.
Jacs Au, 4, 2024
|
|
5V7U
 
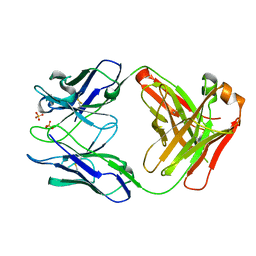 | |
8AWL
 
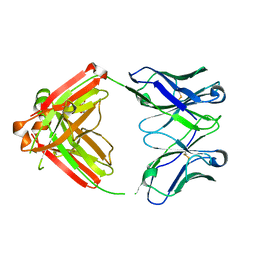 | | Fab RVFV-268 | | Descriptor: | CITRIC ACID, Heavy chain Fab268, Light chain Fab268 | | Authors: | Hulswit, R.J.G, Bowden, T.A, Stass, R. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Multifunctional human monoclonal antibody combination mediates protection against Rift Valley fever virus at low doses.
Nat Commun, 14, 2023
|
|
8AWM
 
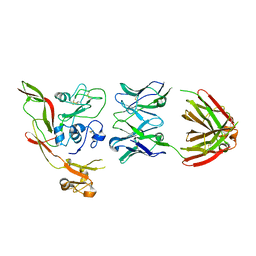 | | RVFV GnH with Fab268 bound | | Descriptor: | Glycoprotein, Heavy chain Fab268, Light chain Fab268 | | Authors: | Hulswit, R.J.G, Bowden, T.A, Stass, R. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multifunctional human monoclonal antibody combination mediates protection against Rift Valley fever virus at low doses.
Nat Commun, 14, 2023
|
|
8AN0
 
 | |
8AH3
 
 | |
