6M8Z
 
 | | Crystal structure of human DJ-1 without a modification on Cys-106 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Shumilin, I.A, Shabalin, I.G, Shumilina, S.V, Werenskjold, C, Utepbergenov, D, Minor, W. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6M81
 
 | | Crystal structure of TylM1 Y14F bound to SAH and dTDP-phenol | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, S-ADENOSYL-L-HOMOCYSTEINE, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
6M83
 
 | | Crystal structure of TylM1 S120A bound to SAH and dTDP-phenol | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3685 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
1R2K
 
 | | Crystal structure of MoaB from Escherichia coli | | Descriptor: | Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Bader, G, Gomez-Ortiz, M, Haussmann, C, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2003-09-28 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the molybdenum-cofactor biosynthesis protein MoaB of Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VGC
 
 | | GAMMA-CHYMOTRYPSIN L-PARA-CHLORO-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | GAMMA CHYMOTRYPSIN, L-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
4ZLO
 
 | | Serine/threonine-protein kinase PAK1 complexed with a dibenzodiazepine: identification of an allosteric site on PAK1 | | Descriptor: | 2,8-difluoro-11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine, GLYCEROL, Serine/threonine-protein kinase PAK 1 | | Authors: | Bellamacina, C.R, Bussiere, D.E. | | Deposit date: | 2015-05-01 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor.
Acs Med.Chem.Lett., 6, 2015
|
|
4ZT9
 
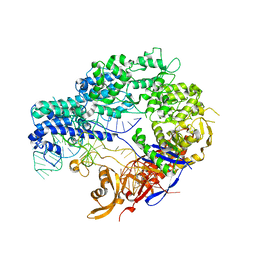 | |
4ZT0
 
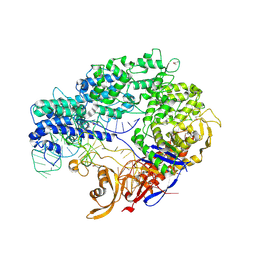 | |
5A0F
 
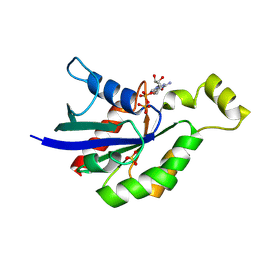 | | Crystal structure of Yersinia Afp18-modified RhoA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Jank, T, Schimpl, M, van Aalten, D.M. | | Deposit date: | 2015-04-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tyrosine glycosylation of Rho by Yersinia toxin impairs blastomere cell behaviour in zebrafish embryos.
Nat Commun, 6, 2015
|
|
5A4C
 
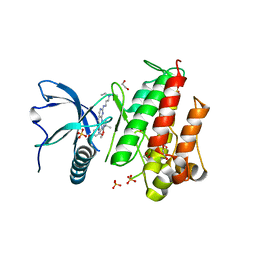 | | FGFR1 ligand complex | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-[2-[3-(diethylamino)propylamino]-6-(3,5-dimethoxyphenyl)pyrido[2,3-d]pyrimidin-7-yl]urea, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
7RYQ
 
 | | Cryo-EM map of KIFBP | | Descriptor: | KIF-binding protein | | Authors: | Tan, Z, Solon, A.L, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7RYP
 
 | | Cryo-EM structure of KIFBP:KIF15 | | Descriptor: | KIF-binding protein, Kinesin-like protein KIF15 | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7D29
 
 | | CBM32 of AlyQ | | Descriptor: | AlyQ, CALCIUM ION | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7S0P
 
 | | Crystal structure of Porcine Factor VIII C2 Domain Bound to Phosphatidylserine | | Descriptor: | Coagulation factor VIII, PHOSPHOSERINE | | Authors: | Peters, S.C, Childers, K.C, Wo, S.W, Brison, C.M, Swanson, C.D, Spiegel, P.C. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Stable binding to phosphatidylserine-containing membranes requires conserved arginine residues in tandem C domains of blood coagulation factor VIII.
Front Mol Biosci, 9, 2022
|
|
3VGN
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 3-fluoro-4-nitrophenol | | Descriptor: | 3-fluoro-4-nitrophenol, Steroid Delta-isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P.A. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VUA
 
 | | Apo IsdH-NEAT3 in space group P3121 at a resolution of 1.85 A | | Descriptor: | ACETATE ION, GLYCEROL, Iron-regulated surface determinant protein H, ... | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-06-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin
To be Published
|
|
6RCX
 
 | | Mycobacterial 4'-phosphopantetheinyl transferase PptAb in complex with the ACP domain of PpsC. | | Descriptor: | CACODYLATE ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
3VIU
 
 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6RA0
 
 | |
6RML
 
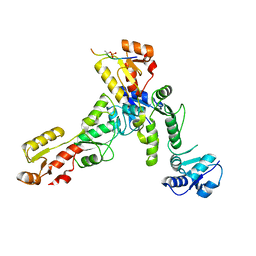 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | Descriptor: | 53BP1, DNA topoisomerase 2-binding protein 1 | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
6RS2
 
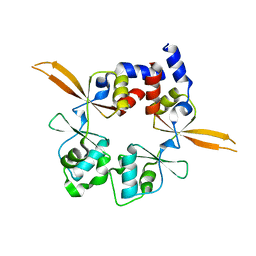 | | Structure of the Bateman module of human CNNM4. | | Descriptor: | Metal transporter CNNM4 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Gomez-Garcia, I, Oyenarte, I, Gimenez, P, Ereno-Orbea, J, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2019-05-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.694 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
3VU4
 
 | |
6RXG
 
 | |
6RNS
 
 | |
6RXF
 
 | |
