5DOZ
 
 | | Crystal structure of JamJ enoyl reductase (NADPH bound) | | Descriptor: | ACETATE ION, JamJ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
5DTD
 
 | |
6KOE
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5DOV
 
 | |
6DEF
 
 | | Vps1 GTPase-BSE fusion complexed with GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Vps1 GTPase-BSE | | Authors: | Ford, M.G.J, Varlakhanova, N.V, Brady, T.M, Chappie, J.S, Hosford, C.J. | | Deposit date: | 2018-05-11 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DWX
 
 | | Crystal structure of SeMet phased viral OTU domain protease from Qalyub virus | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Dzimianski, J.V, Beldon, B.S, Daczkowski, C.M, Goodwin, O.Y, Pegan, S.D. | | Deposit date: | 2018-06-28 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Probing the impact of nairovirus genomic diversity on viral ovarian tumor domain protease (vOTU) structure and deubiquitinase activity.
PLoS Pathog., 15, 2019
|
|
3ERS
 
 | | Crystal Structure of E. coli Trbp111 | | Descriptor: | tRNA-binding protein ygjH | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-10-03 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Trbp111: a tructure specific tRNA binding protein
Embo J., 19, 2000
|
|
6KOC
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | Descriptor: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5E3L
 
 | |
6KSG
 
 | | Vibrio cholerae Methionine Aminopeptidase in holo form | | Descriptor: | GLYCEROL, Methionine aminopeptidase, NICKEL (II) ION, ... | | Authors: | Pillalamarri, V, Addlagatta, A. | | Deposit date: | 2019-08-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine aminopeptidases with short sequence inserts within the catalytic domain are differentially inhibited: Structural and biochemical studies of three proteins from Vibrio spp.
Eur.J.Med.Chem., 209, 2020
|
|
6KSN
 
 | | Structure of a Zn-bound camelid single domain antibody | | Descriptor: | 1,2-ETHANEDIOL, ICab3, SODIUM ION, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2019-08-24 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isolation and structural characterization of a Zn2+-bound single-domain antibody against NorC, a putative multidrug efflux transporter in bacteria.
J.Biol.Chem., 295, 2020
|
|
5E4X
 
 | | Crystal structure of cpSRP43 chromodomain 3 | | Descriptor: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
6KUZ
 
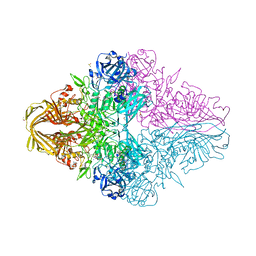 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSL01 | | Descriptor: | 3-(1,3-benzothiazol-2-yl)-2-[[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]methoxy]-5-methyl-benzaldehyde, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Li, X.K, Guo, Y, Li, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated beta-galactosidase.
Chem Sci, 11, 2020
|
|
3HN4
 
 | |
7YLF
 
 | |
6LB8
 
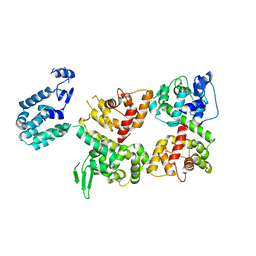 | | Crystal structure of the Ca2+-free T4L-MICU1-MICU2 complex | | Descriptor: | Calcium uptake protein 2, mitochondrial, Endolysin,Calcium uptake protein 1 | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
6LD0
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
7YLG
 
 | |
7YFS
 
 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | Descriptor: | noursin | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
7XY9
 
 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
7P19
 
 | |
6L6L
 
 | | Structural basis of NR4A2 homodimers binding to selective Nur-responsive elements | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*CP*AP*AP*AP*CP*TP*GP*TP*GP*AP*CP*CP*TP*AP*T)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*TP*TP*GP*AP*CP*CP*TP*T)-3'), Nuclear receptor related 1, ... | | Authors: | Jiang, L, Chen, Y. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements.
J.Biol.Chem., 294, 2019
|
|
6KOB
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6KZ0
 
 | | HRV14 3C in complex with single chain antibody GGVV | | Descriptor: | GGVV H chain, GGVV L chain, Genome polyprotein | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
