8PNT
 
 | | Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8PMP
 
 | | Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8PJ6
 
 | | Structure of human 48S translation initiation complex with initiator tRNA, eIF1A and eIF3 (off-pathway) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex.
Nat.Struct.Mol.Biol., 2024
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
6D6V
 
 | | CryoEM structure of Tetrahymena telomerase with telomeric DNA at 4.8 Angstrom resolution | | Descriptor: | DNA (5'-D(P*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*G)-3'), RNA (159-MER), Telomerase associated protein p65, ... | | Authors: | Jiang, J, Wang, Y, Susac, L, Chan, H, Basu, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2018-04-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of Telomerase with Telomeric DNA.
Cell, 173, 2018
|
|
6EM1
 
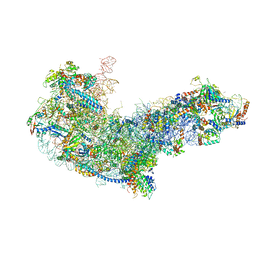 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6ES4
 
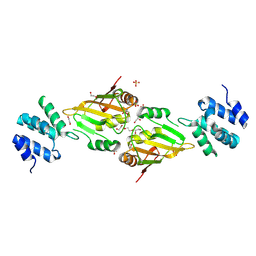 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
6ELZ
 
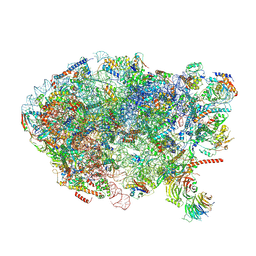 | | State E (TAP-Flag-Ytm1 E80A) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-09-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM5
 
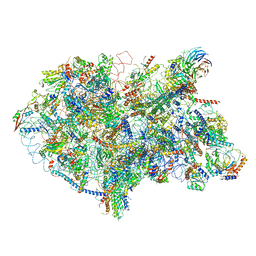 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6F4G
 
 | | 'Crystal structure of the Drosophila melanogaster SNF/U2A'/U2-SL4 complex | | Descriptor: | CHLORIDE ION, Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6F4J
 
 | | Crystal structure of Drosophila melanogaster SNF/U2A' complex | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6EM3
 
 | | State A architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM4
 
 | | State B architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6GD1
 
 | | Structure of HuR RRM3 | | Descriptor: | SODIUM ION, Thioredoxin 1,ELAV-like protein 1 | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6G90
 
 | | Prespliceosome structure provides insight into spliceosome assembly and regulation (map A2) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing factor 39, ... | | Authors: | Plaschka, C, Lin, P.-C, Charenton, C, Nagai, K. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Prespliceosome structure provides insights into spliceosome assembly and regulation.
Nature, 559, 2018
|
|
2MJU
 
 | |
1A9N
 
 | |
2J0S
 
 | | The crystal structure of the Exon Junction Complex at 2.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP *UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
2J0Q
 
 | | The crystal structure of the Exon Junction Complex at 3.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on Mrna.
Cell(Cambridge,Mass.), 126, 2006
|
|
2KT5
 
 | | RRM domain of mRNA export adaptor REF2-I bound to HSV-1 ICP27 peptide | | Descriptor: | ICP27, RNA and export factor-binding protein 2 | | Authors: | Tunnicliffe, N.B, Golovanov, A.P, Wilson, S.A, Hautbergue, G.M. | | Deposit date: | 2010-01-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Cellular mRNA Export Factor REF by Herpes Viral Proteins HSV-1 ICP27 and HVS ORF57.
Plos Pathog., 7, 2011
|
|
2HYI
 
 | | Structure of the human exon junction complex with a trapped DEAD-box helicase bound to RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*U)-3', MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andersen, C.B.F, Le Hir, H, Andersen, G.R. | | Deposit date: | 2006-08-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
2I2Y
 
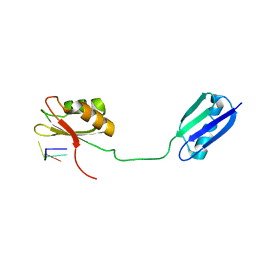 | | Solution structure of the RRM of SRp20 bound to the RNA CAUC | | Descriptor: | (5'-R(*CP*AP*UP*C)-3'), Fusion protein consists of immunoglobulin G-Binding Protein G and Splicing factor, arginine/serine-rich 3 | | Authors: | Hargous, Y.F, Allain, F.H. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Embo J., 25, 2006
|
|
2HGN
 
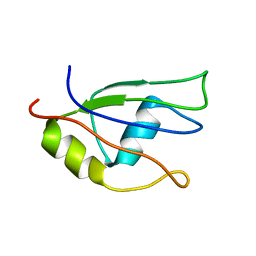 | | NMR structure of the third qRRM domain of human hnRNP F | | Descriptor: | Heterogeneous nuclear ribonucleoprotein F | | Authors: | Dominguez, C, Allain, F.H.-T. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition.
Nucleic Acids Res., 34, 2006
|
|
