1A87
 
 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
1A86
 
 | | MMP8 WITH MALONIC AND ASPARTIC ACID BASED INHIBITOR | | Descriptor: | CALCIUM ION, MMP-8, N-benzyl-N~2~-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-alpha-asparagine, ... | | Authors: | Brandstetter, H, Roedern, E.G.V, Grams, F, Engh, R.A. | | Deposit date: | 1998-04-03 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of malonic acid-based inhibitors bound to human neutrophil collagenase. A new binding mode explains apparently anomalous data.
Protein Sci., 7, 1998
|
|
1PRX
 
 | | HORF6 A NOVEL HUMAN PEROXIDASE ENZYME | | Descriptor: | HORF6 | | Authors: | Choi, H.-J, Kang, S.W, Yang, C.-H, Rhee, S.G, Ryu, S.-E. | | Deposit date: | 1998-04-03 | | Release date: | 1998-06-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
1A88
 
 | | CHLOROPEROXIDASE L | | Descriptor: | CHLOROPEROXIDASE L | | Authors: | Hofmann, B, Toelzer, S, Pelletier, I, Altenbuchner, J, Van Pee, K.-H, Hecht, H.-J. | | Deposit date: | 1998-04-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of the cofactor-free chloroperoxidases.
J.Mol.Biol., 279, 1998
|
|
1URP
 
 | |
1A9C
 
 | | GTP CYCLOHYDROLASE I (C110S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-04-04 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GTP Cyclohydrolase I in Complex with GTP at 2.1 A Resolution
To be Published
|
|
1A9E
 
 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-05 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
1A9I
 
 | |
1SKT
 
 | | SOLUTION STRUCTURE OF APO N-DOMAIN OF TROPONIN C, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
1B5B
 
 | | RAT FERROCYTOCHROME B5 B CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1A9H
 
 | |
1B5A
 
 | | RAT FERROCYTOCHROME B5 A CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1A9G
 
 | |
1A9J
 
 | |
2QWD
 
 | |
1BA4
 
 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
1A9L
 
 | |
1ZAC
 
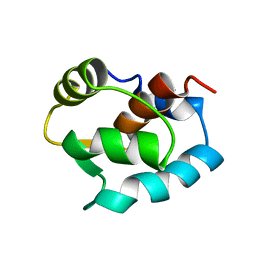 | | N-DOMAIN OF TROPONIN C FROM CHICKEN SKELETAL MUSCLE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
2QWE
 
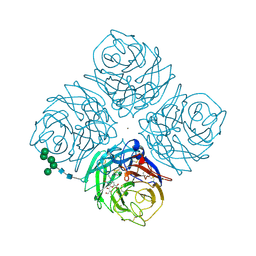 | |
2QWC
 
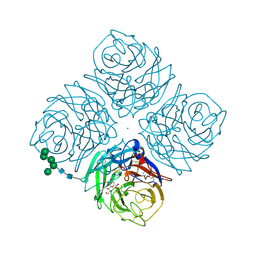 | |
2QWB
 
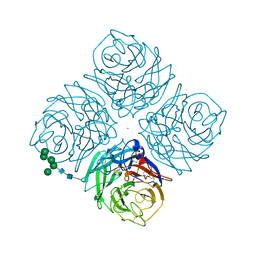 | |
2QWF
 
 | |
2QWK
 
 | |
2A9L
 
 | |
2QWH
 
 | |
