3Q4L
 
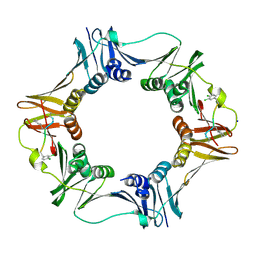 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
1FIB
 
 | |
3CZ2
 
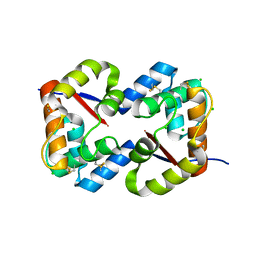 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera at pH 7.0 | | Descriptor: | CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6TPA
 
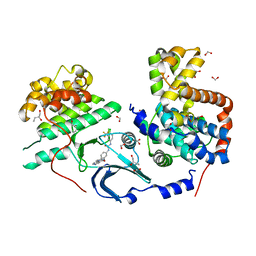 | | CDK8/CyclinC in complex with drug ETP-50775 | | Descriptor: | (2~{R})-butane-1,2-diol, 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-(5-oxidanylidene-6-pyridin-4-yl-pyrido[2,3-b][1,5]benzoxazepin-9-yl)urea, Cyclin-C, ... | | Authors: | Munoz, I.G, Pastor, J, Martinez, S. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-b][1,5]benzoxazepin-5(6H)-one derivatives as CDK8 inhibitors.
Eur.J.Med.Chem., 201, 2020
|
|
1O4U
 
 | |
6VWS
 
 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
3RK3
 
 | | Truncated SNARE complex with complexin | | Descriptor: | Complexin-1, SNAP25, Syntaxin 1a, ... | | Authors: | Kuemmel, D, Reinisch, K.M. | | Deposit date: | 2011-04-17 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Complexin cross-links prefusion SNAREs into a zigzag array.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
2AID
 
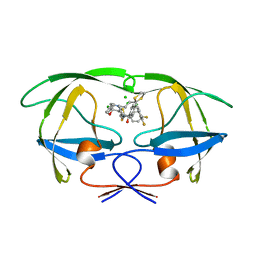 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
6BGH
 
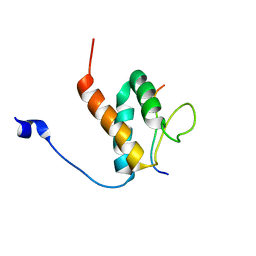 | |
3DLE
 
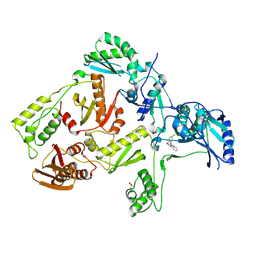 | |
3DM2
 
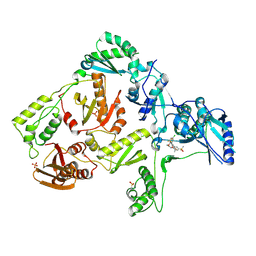 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
2BRQ
 
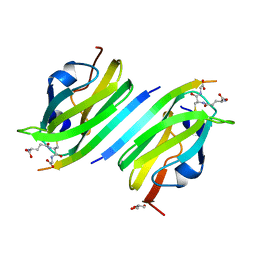 | |
2EEY
 
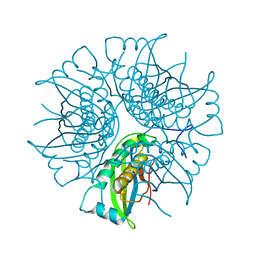 | |
3D77
 
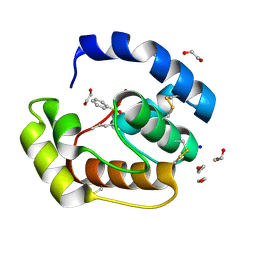 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 4.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3D78
 
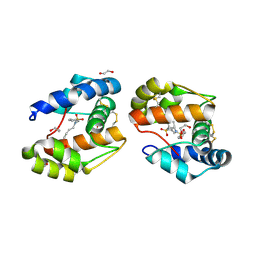 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7KN8
 
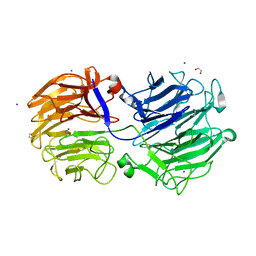 | | Crystal structure of the GH74 xyloglucanase from Xanthomonas campestris (Xcc1752) | | Descriptor: | 1,2-ETHANEDIOL, Cellulase, IODIDE ION, ... | | Authors: | Araujo, E.A, Vieira, P.S, Murakami, M.T, Polikarpov, I. | | Deposit date: | 2020-11-04 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors
Nature Communications, 12, 2021
|
|
1MXT
 
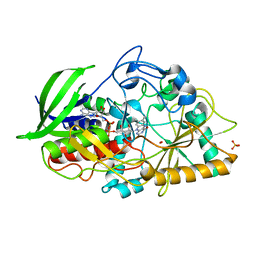 | | Atomic resolution structure of Cholesterol oxidase (Streptomyces sp. SA-COO) | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, OXYGEN MOLECULE, ... | | Authors: | Vrielink, A, Lario, P.I. | | Deposit date: | 2002-10-03 | | Release date: | 2003-02-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sub-atomic resolution crystal structure of cholesterol oxidase:
What atomic resolution crystallography reveals about enzyme mechanism and the role of FAD cofactor in redox activity
J.Mol.Biol., 326, 2003
|
|
3FT5
 
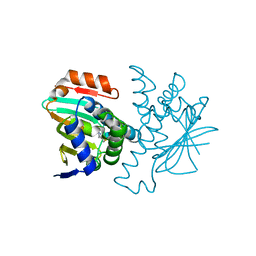 | | Structure of HSP90 bound with a novel fragment | | Descriptor: | 4-methyl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Identification of Hsp90 Inhibitors.
Chemmedchem, 4, 2009
|
|
3FT8
 
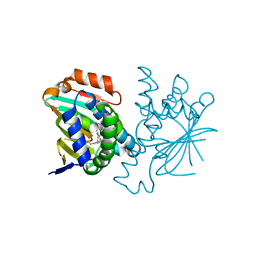 | | Structure of HSP90 bound with a noval fragment. | | Descriptor: | (5E,7S)-2-amino-7-(4-fluoro-2-pyridin-3-ylphenyl)-4-methyl-7,8-dihydroquinazolin-5(6H)-one oxime, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based identification of Hsp90 inhibitors.
Chemmedchem, 4, 2009
|
|
5OBU
 
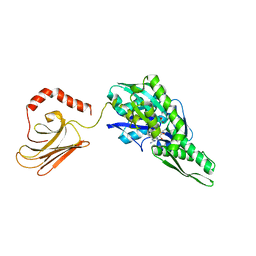 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with AMPPNP. | | Descriptor: | Chaperone protein DnaK, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
3FHT
 
 | |
7L5A
 
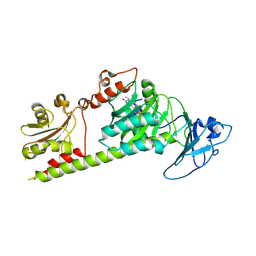 | | Crystal structure of the photosensory module from Xanthomonas campestris bacteriophytochrome XccBphP in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7L59
 
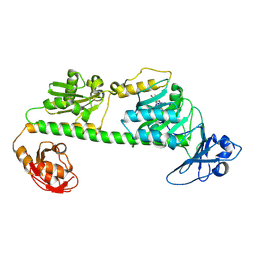 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
