1KDH
 
 | | Binary Complex of Murine Terminal Deoxynucleotidyl Transferase with a Primer Single Stranded DNA | | Descriptor: | 5'-D(P*(BRU)P*(BRU)P*(BRU)P*(BRU))-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
8AHH
 
 | |
8AHG
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHI
 
 | |
2DYW
 
 | | A Backbone binding DNA complex | | Descriptor: | (6-AMINOHEXYLAMINE)(TRIAMMINE) PLATINUM(II) COMPLEX, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', SODIUM ION, ... | | Authors: | Komeda, S, Moulaei, T, Woods, K.K, Chikuma, M, Farrell, N.P, Williams, L.D. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A Third Mode of DNA Binding: Phosphate Clamps by a Polynuclear Platinum Complex
J.Am.Chem.Soc., 128, 2006
|
|
1RRS
 
 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1RRQ
 
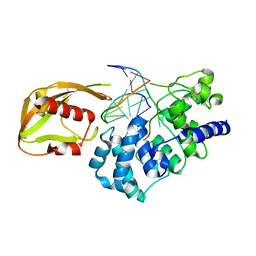 | | MutY adenine glycosylase in complex with DNA containing an A:oxoG pair | | Descriptor: | 5'-D(*TP*GP*TP*CP*CP*AP*AP*GP*TP*CP*T)-3', 5'-D(AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
8DW7
 
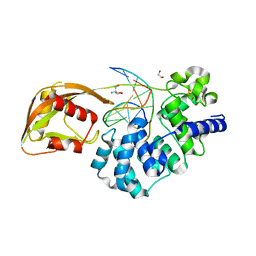 | | DNA glycosylase MutY variant N146S in complex with DNA containing the transition state analog 1N paired with d(8-oxo-G) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-31 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DVY
 
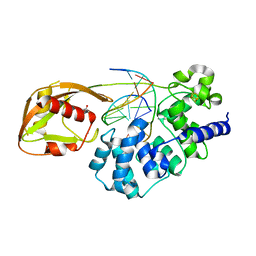 | | DNA glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site product (AP) and crystalized with calcium acetate | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
1GV6
 
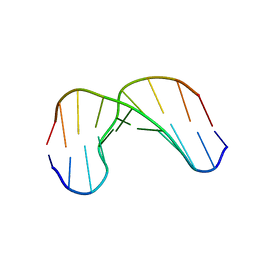 | | Solution structure of alfa-L-LNA:DNA duplex | | Descriptor: | 5- D(*CP*(ATL)P*GP*CP*(ATL)P*(ATL)P*CP*(ATL)P* GP*C) -3, 5- D(*GP*CP*AP*GP*AP*AP*GP*CP*AP*G) -3 | | Authors: | Nielsen, K.M.E, Petersen, M, Haakansson, A.E, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alfa-L-Lna (Alfa-L-Ribo Configured Locked Nucleic Acid) Recognition of DNA: An NMR Spectroscopic Study
Chemistry, 8, 2002
|
|
8DWD
 
 | | Adenine glycosylase MutY variant E43S in complex with DNA containing d(8-oxo-G) paired with an AP site generated by the enzyme acting on purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
8DWF
 
 | | Glycosylase MutY variant E43S in complex with DNA containing d(8-oxo-G) paired with substrate adenine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
8DWE
 
 | | Adenine glycosylase MutY variant E43Q in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
7XX6
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
1QTW
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1VRL
 
 | | MutY adenine glycosylase in complex with DNA and soaked adenine free base | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', ADENINE, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2005-03-08 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1NGN
 
 | | Mismatch repair in methylated DNA. Structure of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4 | | Descriptor: | methyl-CpG binding protein MBD4 | | Authors: | Wu, P, Qiu, C, Sohail, A, Zhang, X, Bhagwat, A.S, Cheng, X. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4
J.Biol.Chem., 278, 2003
|
|
2BGT
 
 | |
7YVW
 
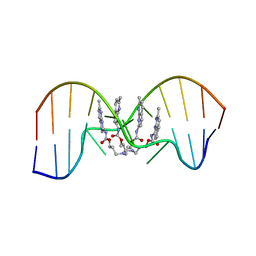 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
2BGU
 
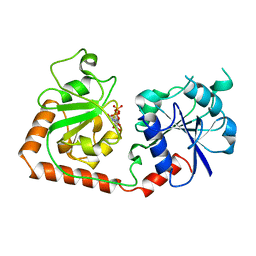 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
7UIN
 
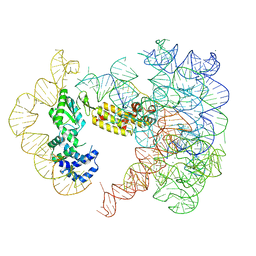 | | CryoEM Structure of an Group II Intron Retroelement | | Descriptor: | AMMONIUM ION, DNA (37-MER), E.r IIC Intron, ... | | Authors: | Chung, K, Xu, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a mobile intron retroelement poised to attack its structured DNA target
Science, 378, 2022
|
|
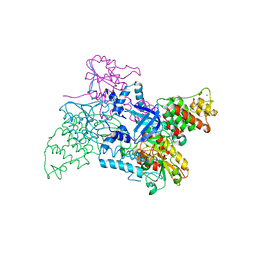
2V1C
 
 | |
2IRU
 
 | |
