3S6L
 
 | |
4I71
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with a trypanocidal compound | | Descriptor: | (2R,3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-2-(hydroxymethyl)pyrrolidine-3,4-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Giannese, F, Degano, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3S73
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S76
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1H-imidazole-2-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4I7E
 
 | | Crystal Structure of the Bacillus stearothermophilus Phosphofructokinase Mutant D12A in Complex with PEP | | Descriptor: | 6-phosphofructokinase, PHOSPHOENOLPYRUVATE | | Authors: | Mosser, R, Reddy, M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redefining the Role of the Quaternary Shift in Bacillus stearothermophilus Phosphofructokinase.
Biochemistry, 52, 2013
|
|
4I8K
 
 | | Bovine trypsin at 0.85 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2NQD
 
 | | Crystal structure of cysteine protease inhibitor, chagasin, in complex with human cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin L, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-31 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
4HJK
 
 | | U7Ub7 Disulfide variant | | Descriptor: | PHOSPHATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
3S8X
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(4-methyl-6-oxo-1,6-dihydro-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 4-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-05-31 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
4I94
 
 | | Structure of BSK8 in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable serine/threonine-protein kinase At5g41260 | | Authors: | Gruetter, C, Rauh, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-08-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of the RLCK Family Member BSK8: A Pseudokinase with an Unprecedented Architecture
J.Mol.Biol., 425, 2013
|
|
3S8Q
 
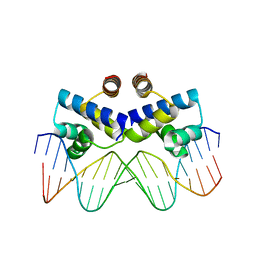 | | Crystal structure of the R-M controller protein C.Esp1396I OL operator complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*AP*CP*TP*TP*AP*TP*AP*GP*TP*CP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*GP*AP*CP*TP*AP*TP*AP*AP*GP*TP*CP*AP*CP*A)-3'), R-M CONTROLLER PROTEIN | | Authors: | McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-05-30 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex.
Nucleic Acids Res., 40, 2012
|
|
4DG1
 
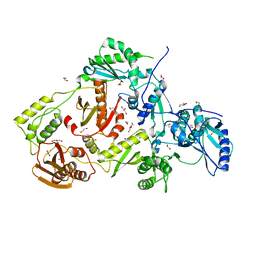 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
4DLU
 
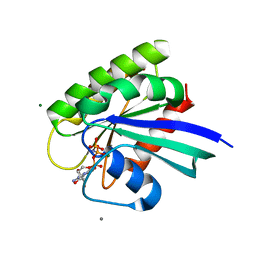 | | H-Ras Set 1 Ca(OAc)2, on | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
4DGQ
 
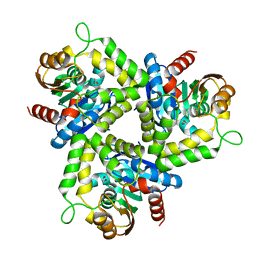 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
3SAP
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(5-butyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-{[(5-butylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
4DIX
 
 | |
3SAX
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-{[(5-ethyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-chloro-5-{[(5-ethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
4HLE
 
 | | Compound 21 (1-alkyl-substituted 1,2,4-triazoles) | | Descriptor: | 2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepine-8-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Rouge, L, Wu, P. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Cis-Amide isosteric replacement in thienobenzoxepin inhibitors of PI3-kinase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3SB9
 
 | | Cu-mediated Dimer of T4 Lysozyme R76H/R80H by Synthetic Symmetrization | | Descriptor: | COPPER (II) ION, FORMIC ACID, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SBX
 
 | |
3SBY
 
 | | Crystal Structure of SeMet-Substituted Apo-MMACHC (1-244), a human B12 processing enzyme | | Descriptor: | Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
3SDO
 
 | |
4DQ6
 
 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Putative pyridoxal phosphate-dependent transferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
3SDW
 
 | |
4DO4
 
 | | Pharmacological chaperones for human alpha-N-acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2012-02-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pharmacological chaperones for human alpha-N-acetylgalactosaminidase
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
