3N8E
 
 | | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin) | | Descriptor: | Stress-70 protein, mitochondrial | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin)
To be published
|
|
8ERM
 
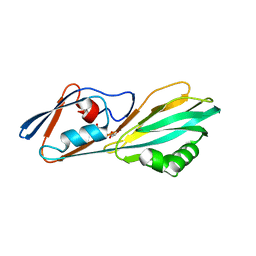 | | Crystal structure of FliC D2/D3 domains from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellin, GLYCEROL, SULFATE ION | | Authors: | Nedeljkovic, M, Bonsor, D.A, Postel, S, Sundberg, E.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments.
Plos Pathog., 19, 2023
|
|
2O2N
 
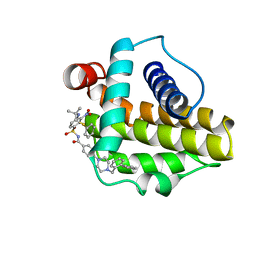 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-[4-(BIPHENYL-2-YLMETHYL)PIPERAZIN-1-YL]-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
1H8C
 
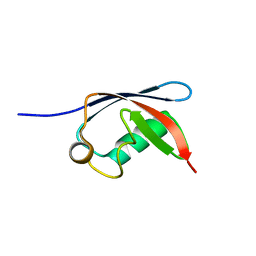 | | UBX domain from human faf1 | | Descriptor: | FAS-ASSOCIATED FACTOR 1 | | Authors: | Bycroft, M.M. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ubx Domain: A Widespread Ubiquitin-Like Module
J.Mol.Biol., 307, 2001
|
|
5EQU
 
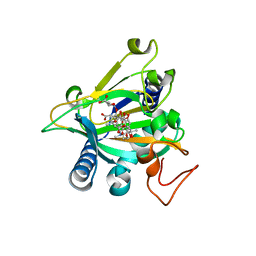 | | Crystal structure of the epimerase SnoN in complex with Fe3+, alpha ketoglutarate and nogalamycin RO | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Nogalamycin RO, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-13 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2NUT
 
 | |
8DDG
 
 | |
2PQ9
 
 | | E. coli EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
5EPA
 
 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2OMQ
 
 | |
2OL3
 
 | | crystal structure of BM3.3 ScFV TCR in complex with PBM8-H-2KBM8 MHC class I molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALLOGENEIC H-2KBM8 MHC CLASS I MOLECULE, BM3.3 T-CELL RECEPTOR ALPHA-CHAIN, ... | | Authors: | Mazza, C, Auphan-Anezin, N, Gregoire, C, Guimezanes, A, Kellenberger, C, Roussel, A, Kearney, A, Van der Merwe, P.A, Schmitt-Verhulst, A.M, Malissen, B. | | Deposit date: | 2007-01-18 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides?
Embo J., 26, 2007
|
|
7B34
 
 | | MST3 in complex with compound MRIA12 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranyl-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
2NVQ
 
 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with 2'dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NXE
 
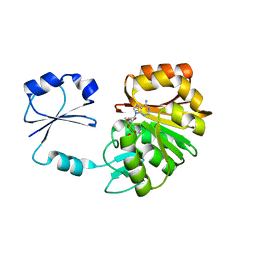 | | T. thermophilus ribosomal protein L11 methyltransferase (PrmA) in complex with S-Adenosyl-L-Methionine | | Descriptor: | Ribosomal protein L11 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA.
Embo J., 26, 2007
|
|
2O39
 
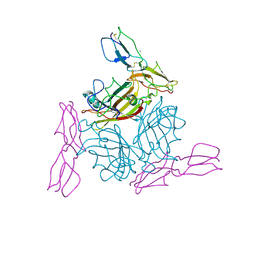 | | Human Adenovirus type 11 knob in complex with domains SCR1 and SCR2 of CD46 (membrane cofactor protein, MCP) | | Descriptor: | CALCIUM ION, Fiber protein, Membrane cofactor protein, ... | | Authors: | Persson, D.B, Reiter, D.M, Arnberg, N, Stehle, T. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Adenovirus type 11 binding alters the conformation of its receptor CD46.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2NVX
 
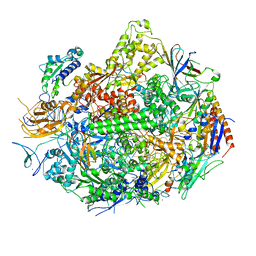 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
6ANY
 
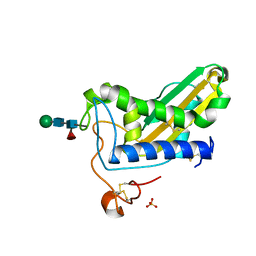 | | Structure of BmVAL-1 | | Descriptor: | Bm4233, isoform b, SULFATE ION, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2017-08-14 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Brugia malayi venom allergen-like protein-1 (BmVAL-1), a vaccine candidate for lymphatic filariasis.
Int. J. Parasitol., 48, 2018
|
|
2PQ5
 
 | | Crystal structure of Dual specificity protein phosphatase 13 (DUSP13) | | Descriptor: | Dual specificity protein phosphatase 13 | | Authors: | Ugochukwu, E, Salah, E, Savitsky, P, Barr, A, Pantic, N, Niesen, F, Burgess-Brown, N, Berridge, G, Bunkoczi, G, Uppenberg, J, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dual specificity protein phosphatase 13 (DUSP13).
To be Published
|
|
6GGG
 
 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | 2-[(3~{S})-7-fluoranyl-6-(2-methylpropyl)-4-[(3-oxidanylidene-4~{H}-1,4-benzoxazin-6-yl)carbonyl]-2,3-dihydro-1,4-benzoxazin-3-yl]-~{N}-methyl-ethanamide, CHLORIDE ION, Mineralocorticoid receptor, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
2PIS
 
 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
5JSB
 
 | | Crystal structure of Mcl1-inhibitor complex | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 inhibitor | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2016-05-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Elife, 5, 2016
|
|
2B8F
 
 | |
7B33
 
 | | MST3 in complex with MRIA11 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[4-[6-[bis(fluoranyl)methyl]pyridin-2-yl]-2-chloranyl-phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
2Q3H
 
 | | The crystal structure of RhouA in the GDP-bound state. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras homolog gene family, ... | | Authors: | Gileadi, C, Yang, X, Papagrigoriou, E, Elkins, J, Zhao, Y, Bray, J, Gileadi, O, Umeano, C, Ugochukwu, E, Uppenberg, J, Bunkoczi, G, von Delft, F, Pike, A.C.W, Phillips, C, Savitsky, P, Fedorov, O, Edwards, A, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of RhouA in the GDP-bound state.
To be Published
|
|
7B30
 
 | | MST3 in complex with compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
