5UQ7
 
 | | 70S ribosome complex with dnaX mRNA stemloop and E-site tRNA ("in" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
5QCM
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[[(1~{S})-2-[(~{E})-3-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]phenyl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCL
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
6DL7
 
 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
3D3C
 
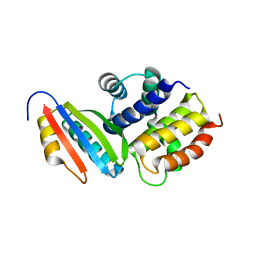 | |
5QCN
 
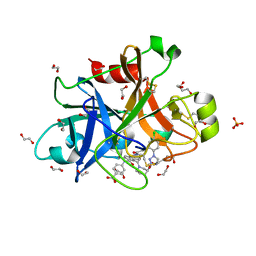 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5FMG
 
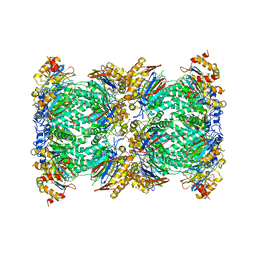 | | Structure and function based design of Plasmodium-selective proteasome inhibitors | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, BETA3 PROTEASOME SUBUNIT, PUTATIVE, ... | | Authors: | Li, H, O'Donoghue, A.J, van der Linden, W.A, Xie, S.C, Yoo, E, Foe, I.T, Tilley, L, Craik, C.S, da Fonseca, P.C.A, Bogyo, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-02 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Function Based Design of Plasmodium-Selective Proteasome Inhibitors
Nature, 530, 2016
|
|
3D3B
 
 | |
6HWU
 
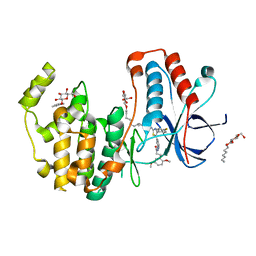 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azothiazol-based Inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
8ATY
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with JP85 (compound 1). | | Descriptor: | 2-[4-chloranyl-6-(5,6,7,8-tetrahydronaphthalen-1-ylamino)pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
3DHX
 
 | | Crystal structure of isolated C2 domain of the methionine uptake transporter | | Descriptor: | IODIDE ION, Methionine import ATP-binding protein metN | | Authors: | Johnson, E, Kaiser, J.T, Lee, A.T, Rees, D.C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
5UQ8
 
 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-03-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
5I12
 
 | | Crystal structure of the catalytic domain of MMP-9 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-05 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
3DW7
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
7A1H
 
 | | Crystal structure of wild-type CI2 | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
7A3M
 
 | |
6D3Q
 
 | | Crystal structure of Escherichia coli enolase complexed with a natural inhibitor SF2312. | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Erlandsen, H, Krucinska, J, Hazeen, A, Wright, D. | | Deposit date: | 2018-04-16 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6GS1
 
 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS7
 
 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
3DW4
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DVZ
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
1S1U
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
5D7J
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64(Y95alphaF) TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
2NYB
 
 | |
