4FJ9
 
 | | RB69 DNA polymerase ternary complex with dTTP/dT | | Descriptor: | CALCIUM ION, DNA polymerase, DNA primer, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FJX
 
 | | RB69 DNA polymerase ternary complex with dATP/dG | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-12 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FJG
 
 | | RB69 DNA polymerase ternary complex with dATP/dC | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FK0
 
 | | RB69 DNA polymerase ternary complex with dCTP/dG | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-12 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FJ8
 
 | | RB69 DNA polymerase ternary complex with dCTP/dT | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FJ5
 
 | | RB69 DNA polymerase ternary complex with dATP/dT | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FJK
 
 | | RB69 DNA polymerase ternary complex with dATP/dA | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
4FJM
 
 | | RB69 DNA polymerase ternary complex with dCTP/dA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
7VW3
 
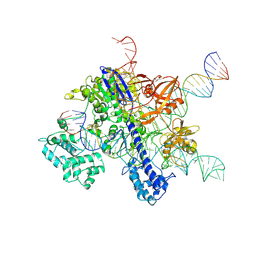 | | Cryo-EM structure of SaCas9-sgRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, MAGNESIUM ION, Non-target DNA strand, ... | | Authors: | Du, W.H, Qian, J.Q, Huang, Q. | | Deposit date: | 2021-11-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Full-Length Model of SaCas9-sgRNA-DNA Complex in Cleavage State.
Int J Mol Sci, 24, 2023
|
|
3M1M
 
 | | Crystal structure of the primase-polymerase from Sulfolobus islandicus | | Descriptor: | GLYCEROL, ORF904, SULFATE ION, ... | | Authors: | Vannini, A, Beck, K, Lipps, G, Cramer, P. | | Deposit date: | 2010-03-05 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis
Nucleic Acids Res., 38, 2010
|
|
6N8K
 
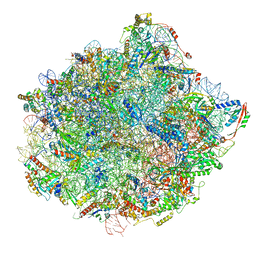 | | Cryo-EM structure of early cytoplasmic-immediate (ECI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
2JQV
 
 | |
6P0K
 
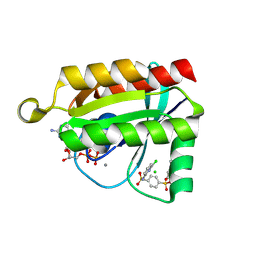 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(6-chloropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4V
 
 | |
6P0N
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(6-fluoropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0M
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(3-chloropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0I
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5-methoxypyridin-3-yl)sulfamoyl]benzene-1-sulfonic acid, ACETATE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0L
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(2-methoxypyridin-3-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4F
 
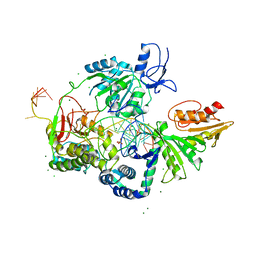 | | Crystal structure of the XPB-Bax1-forked DNA ternary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*TP*TP*TP*GP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*GP*TP*AP*GP*GP*TP*TP*TP*CP*CP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | He, F, Hilario, E, Fan, L. | | Deposit date: | 2019-05-27 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of the XPB helicase-Bax1 nuclease complex interacting with the repair bubble DNA.
Nucleic Acids Res., 48, 2020
|
|
6T9V
 
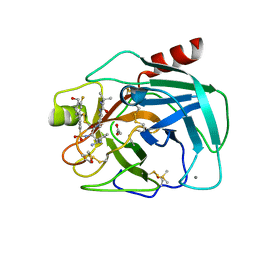 | | Bovine Trypsin in complex with the synthetic inhibitor (S)-3-(3-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-2-((3'-fluoro-4'-(hydroxymethyl)-[1,1'-biphenyl])-3-sulfonamido)-3-oxopropyl)benzimidamide (MI-1904) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-[3-fluoranyl-4-(hydroxymethyl)phenyl]phenyl]sulfonylamino ]propanoyl]piperidin-4-yl]urea, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Merkl, S, Keils, A, Mueller, J.M, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12642729 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
7BHD
 
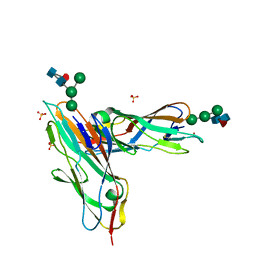 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
4FJ1
 
 | | Crystal Structure of the ternary complex between a fungal 17beta-hydroxysteroid dehydrogenase (Holo form) and genistein | | Descriptor: | 17beta-hydroxysteroid dehydrogenase, DIMETHYL SULFOXIDE, GENISTEIN, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I. | | Deposit date: | 2012-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
6T9T
 
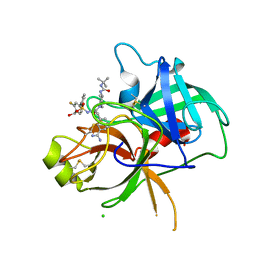 | | Matriptase in complex with the synthetic inhibitor (S)-3-(3-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-2-((3'-fluoro-4'-(hydroxymethyl)-[1,1'-biphenyl])-3-sulfonamido)-3-oxopropyl)benzimidamide (MI-1904) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-[3-fluoranyl-4-(hydroxymethyl)phenyl]phenyl]sulfonylamino ]propanoyl]piperidin-4-yl]urea, CHLORIDE ION, Suppressor of tumorigenicity 14 protein | | Authors: | Mueller, J.M, Merkl, S, Keils, A, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
2KY5
 
 | | Solution structure of the PECAM-1 cytoplasmic tail with DPC | | Descriptor: | Platelet endothelial cell adhesion molecule | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Paddock, C, Newman, D.K, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Residues within a lipid-associated segment of the PECAM-1 cytoplasmic domain are susceptible to inducible, sequential phosphorylation.
Blood, 117, 2011
|
|
2KPV
 
 | |
