7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-28 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
2IVH
 
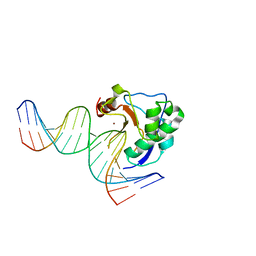 | | Crystal structure of the nuclease domain of ColE7 (H545Q mutant) in complex with an 18-bp duplex DNA | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*TP*CP*GP*AP*TP *CP*GP*AP*AP*TP*TP*CP*C)-3', COLCIN-E7, ZINC ION | | Authors: | Wang, Y.T, Yang, W.J, Li, C.L, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2006-06-13 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Sequence-Dependent DNA Cleavage by Nonspecific Endonucleases.
Nucleic Acids Res., 35, 2007
|
|
2J2M
 
 | | Crystal Structure Analysis of Catalase from Exiguobacterium oxidotolerans | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hara, I, Ichise, N, Kojima, K, Kondo, H, Ohgiya, S, Matsuyama, H, Yumoto, I. | | Deposit date: | 2006-08-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Relationship between the Size of the Bottleneck 15 a from Iron in the Main Channel and the Reactivity of Catalase Corresponding to the Molecular Size of Substrates.
Biochemistry, 46, 2007
|
|
2IVK
 
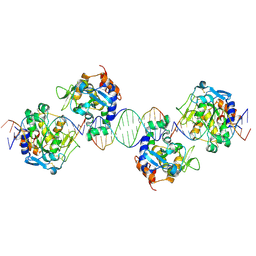 | | Crystal structure of the periplasmic endonuclease Vvn complexed with a 16-bp DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*CP*GP*AP*TP*CP*GP *AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*CP*GP*AP*TP*CP *GP*AP*AP*TP*T)-3', 5'-D(*GP*AP*AP*TP*TP*CP*GP*AP*TP*CP *GP*AP*AP*TP*TP*C)-3', ... | | Authors: | Wang, Y.T, Yang, W.J, Li, C.L, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2006-06-14 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Sequence-Dependent DNA Cleavage by Nonspecific Endonucleases.
Nucleic Acids Res., 35, 2007
|
|
4XZN
 
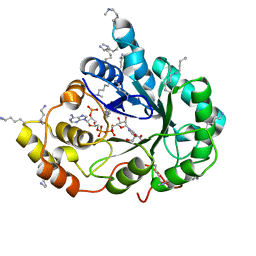 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZL
 
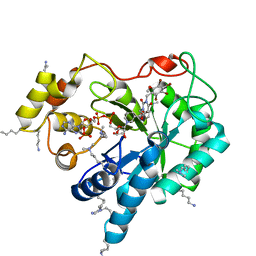 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZM
 
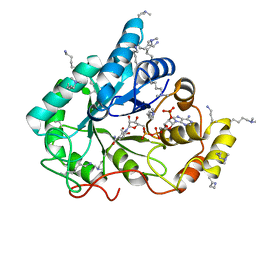 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
7OSA
 
 | | Pre-translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
7OSM
 
 | | Intermediate translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
7OII
 
 | | CspA-70 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OFQ
 
 | |
7OIF
 
 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OLC
 
 | | Thermophilic eukaryotic 80S ribosome at idle POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
4WOI
 
 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
4YBB
 
 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7OW7
 
 | |
2IHS
 
 | |
4WF1
 
 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2LR9
 
 | |
1IW7
 
 | |
2NLI
 
 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
2O4C
 
 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
