7YQ2
 
 | | Crystal structure of photosystem II expressing psbA2 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
4HMX
 
 | | Crystal structure of PhzG from Burkholderia lata 383 in complex with tetrahydrophenazine-1-carboxylic acid | | Descriptor: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase | | Authors: | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7YQ7
 
 | | Crystal structure of photosystem II expressing psbA3 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
3TZQ
 
 | |
2PHD
 
 | | Crystal Structure Determination of a Salicylate 1,2-Dioxygenase from Pseudaminobacter salicylatoxidans | | Descriptor: | ACETATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F. | | Deposit date: | 2007-04-11 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure Determination of a Salicylate 1,2-Dioxygenase from Pseudaminobacter salicylatoxidans
To be Published
|
|
3ZG3
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH THE PYRIDINE INHIBITOR N-(1-(5-(trifluoromethyl)(pyridin-2-yl)) piperidin-4yl)-N-(4-(trifluoromethyl)phenyl)pyridin-3-amine (EPL- BS967, UDD) | | Descriptor: | N-[4-(trifluoromethyl)phenyl]-N-[1-[5-(trifluoromethyl)pyridin-2-yl]piperidin-4-yl]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
1PBE
 
 | |
1HZS
 
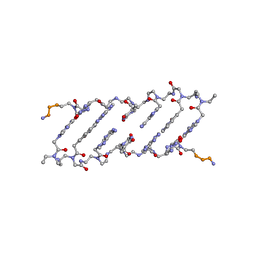 | | Crystal structure of a peptide nucleic acid duplex (BT-PNA) containing a bicyclic analogue of thymine | | Descriptor: | PEPTIDE NUCLEIC ACID | | Authors: | Eldrup, A.B, Nielsen, B.B, Haaima, G, Rasmussen, H, Kastrup, J.S, Christensen, C, Nielsen, P.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,8-Naphthyridin-2(1H)-ones. Novel Bicyclic and Tricyclic Analogues of Thymine in Peptide Nucleic Acids (PNAs)
Eur.J.Org.Chem., 9, 2001
|
|
5WS5
 
 | | Native XFEL structure of photosystem II (preflash dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
4IMT
 
 | | Structure of rat neuronal nitric oxide synthase in complex with 6,6'-((4-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[4-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided design of selective inhibitors of neuronal nitric oxide synthase.
J.Med.Chem., 56, 2013
|
|
3GNC
 
 | |
3R9B
 
 | | Crystal structure of Mycobacterium smegmatis CYP164A2 in ligand free state | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 164A2, DODECANE, ... | | Authors: | Agnew, C.R.J, Warrilow, A.G.S, Kelly, S.L, Brady, R.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | An enlarged, adaptable active site in CYP164 family P450 enzymes, the sole P450 in Mycobacterium leprae.
Antimicrob.Agents Chemother., 56, 2012
|
|
3LD4
 
 | | Urate oxidase complexed with 8-nitro xanthine | | Descriptor: | 1,2-ETHANEDIOL, 8-nitro-3,7-dihydro-1H-purine-2,6-dione, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Gabison, L. | | Deposit date: | 2010-01-12 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2ZI8
 
 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis in complex with 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (DHSA) | | Descriptor: | 3,4-dihydroxy-9,10-secoandrosta-1(10),2,4-triene-9,17-dione, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2008-02-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
3FG1
 
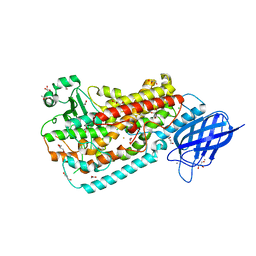 | | Crystal structure of Delta413-417:GS LOX | | Descriptor: | ACETIC ACID, Allene oxide synthase-lipoxygenase protein, CALCIUM ION, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
4N4L
 
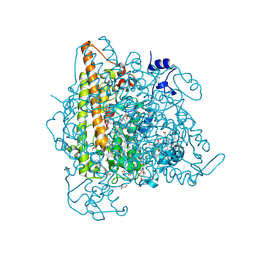 | | Kuenenia stuttgartiensis hydroxylamine oxidoreductase soaked in hydrazine | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-cyclohexylbutanoyl)(2-hydroxyethyl)amino]-1-deoxy-D-glucitol, HEME C, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
4BJK
 
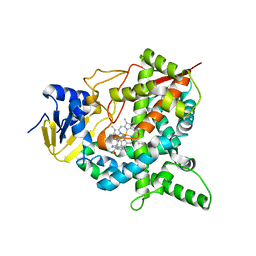 | |
3WEU
 
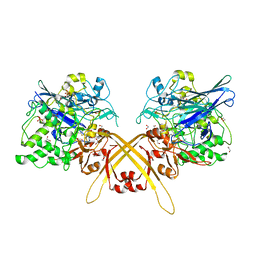 | | Crystal structure of the L-Lys epsilon-oxidase from Marinomonas mediterranea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | Authors: | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
1H69
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
4CK9
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFT) | | Descriptor: | (1S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl [4-(propan-2-yl)phenyl]carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4NHM
 
 | | Crystal structure of Tpa1p from Saccharomyces cerevisiae, termination and polyadenylation protein 1, in complex with N-[(1-chloro-4-hydroxyisoquinolin-3-yl)carbonyl]glycine (IOX3/UN9) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
3FG3
 
 | | Crystal structure of Delta413-417:GS I805W LOX | | Descriptor: | ACETIC ACID, Allene oxide synthase-lipoxygenase protein, CALCIUM ION, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
3WEV
 
 | | Crystal structure of the Schiff base intermediate of L-Lys epsilon-oxidase from Marinomonas mediterranea with L-Lys | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | Authors: | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
5WS6
 
 | | Native XFEL structure of Photosystem II (preflash two-flash dataset | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
1W9M
 
 | | AS-isolated hybrid cluster protein from Desulfovibrio vulgaris X-ray structure at 1.35A resolution using iron anomalous signal | | Descriptor: | FE-S-O HYBRID CLUSTER, HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Coelho, D, Romao, C.V, Teixeira, M, Lindley, P.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Relationships in the Hybrid Cluster Protein Family:Structure of the Anaerobically Purified Hybrid Cluster Protein from Desulfovibrio Vulgaris at 1.35 A Resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
