5GVS
 
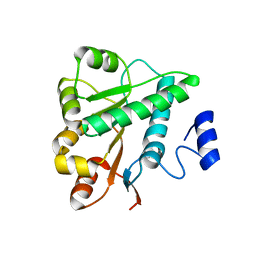 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
5GVR
 
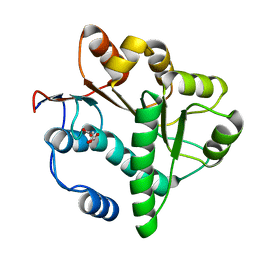 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
8IK3
 
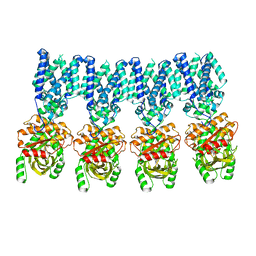 | |
8I6G
 
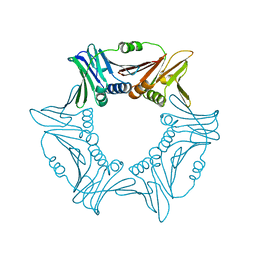 | |
8I6H
 
 | |
6CFF
 
 | | Stimulator of Interferon Genes Human | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
6DNK
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
7F60
 
 | |
2K36
 
 | | Structure ensemble Backbone and side-chain 1H, 13C, and 15N Chemical Shift Assignments, 1H-15N RDCs and 1H-1H nOe restraints for protein K7 from the Vaccinia virus | | Descriptor: | Protein K7 | | Authors: | Kalverda, A.P, Thompson, G.S, Vogel, A, Schr der, M, Bowie, A.G, Khan, A.R, Homans, S.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3.
J.Mol.Biol., 385, 2009
|
|
8P01
 
 | | Crystal structure of human STING ectodomain in complex with BI 7446, a potent cyclic dinucleotide STING agonist with broad-spectrum variant activity for the treatment of cancer | | Descriptor: | 3-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9-fluoranyl-18-oxidanyl-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-6~{H}-imidazo[4,5-d]pyridazin-7-one, Stimulator of interferon genes protein | | Authors: | Nar, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Discovery of BI 7446: A Potent Cyclic Dinucleotide STING Agonist with Broad-Spectrum Variant Activity for the Treatment of Cancer.
J.Med.Chem., 66, 2023
|
|
6CY7
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
8P45
 
 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
4EFO
 
 | | Crystal structure of the ubiquitin-like domain of human TBK1 | | Descriptor: | Serine/threonine-protein kinase TBK1 | | Authors: | Li, J, Li, J, Miyahira, A, Sun, J, Liu, Y, Cheng, G, Liang, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of the ubiquitin-like domain of human TBK1.
Protein Cell, 3, 2012
|
|
4EMT
 
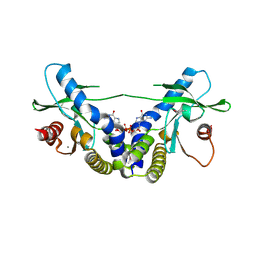 | | Crystal Structure of human STING bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EMU
 
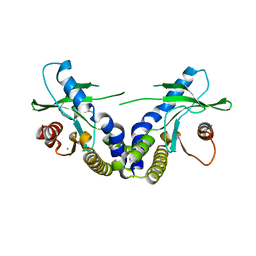 | | Crystal structure of ligand free human STING | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3PHU
 
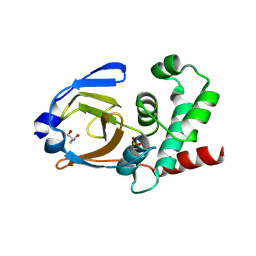 | | OTU Domain of Crimean Congo Hemorrhagic Fever Virus | | Descriptor: | GLYCEROL, RNA-directed RNA polymerase L | | Authors: | Akutsu, M, Ye, Y, Virdee, S, Komander, D. | | Deposit date: | 2010-11-04 | | Release date: | 2011-02-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6X5A
 
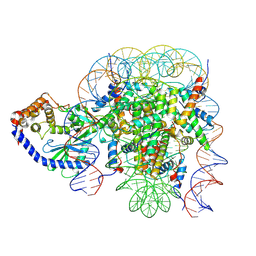 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
3MWP
 
 | | Nucleoprotein structure of lassa fever virus | | Descriptor: | Nucleoprotein, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX5
 
 | | Lassa fever virus nucleoprotein complexed with UTP | | Descriptor: | Nucleoprotein, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
6X59
 
 | | The mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA, Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
3PHX
 
 | | OTU Domain of Crimean Congo Hemorrhagic Fever Virus in complex with ISG15 | | Descriptor: | ACETIC ACID, ETHANAMINE, RNA-directed RNA polymerase L, ... | | Authors: | Akutsu, M, Ye, Y, Virdee, S, Komander, D. | | Deposit date: | 2010-11-04 | | Release date: | 2011-02-02 | | Last modified: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4QXO
 
 | | Crystal structure of hSTING(group2) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
4QXR
 
 | | Crystal structure of hSTING(S162A/G230I/Q266I) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
3PHW
 
 | | OTU Domain of Crimean Congo Hemorrhagic Fever Virus in complex with Ubiquitin | | Descriptor: | ETHANAMINE, RNA-directed RNA polymerase L, Ubiquitin-40S ribosomal protein S27a | | Authors: | Akutsu, M, Ye, Y, Virdee, S, Komander, D. | | Deposit date: | 2010-11-04 | | Release date: | 2011-02-02 | | Last modified: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6XNN
 
 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
