3H4L
 
 | | Crystal Structure of N terminal domain of a DNA repair protein | | Descriptor: | DNA mismatch repair protein PMS1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Arana, M.E, Holmes, S.F, Fortune, J.M, Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2009-04-20 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional residues on the surface of the N-terminal domain of yeast Pms1.
Dna Repair, 9, 2010
|
|
2LIB
 
 | | DNA sequence context conceals alpha anomeric lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*TP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*(A3A)P*GP*GP*AP*CP*G)-3') | | Authors: | Johnson, C.N, Spring, A.M, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2011-08-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA sequence context conceals alpha-anomeric lesions.
J.Mol.Biol., 416, 2012
|
|
7YWH
 
 | | Six DNA Helix Bundle nanopore - State 1 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWL
 
 | | Six DNA Helix Bundle nanopore - State 3 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWI
 
 | | Six DNA duplex bundle nanopore - State 2 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWO
 
 | | Six DNA Helix Bundle nanopore - State 5 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWN
 
 | | Six DNA Helix Bundle nanopore - State 4 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Orlova, E.V, Coveney, P, Howorka, S. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
1DKY
 
 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 2 NATIVE CRYSTALS | | Descriptor: | DNAK, PEPTIDE SUBSTRATE | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
9DNA
 
 | |
1XBL
 
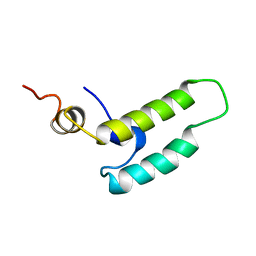 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
1BQZ
 
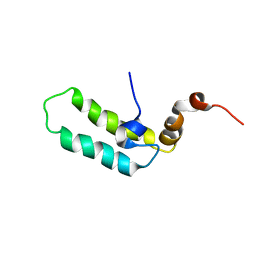 | |
1BPR
 
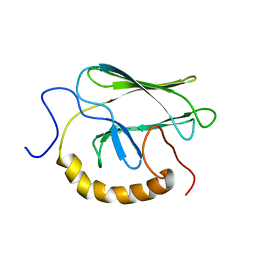 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
1BQ0
 
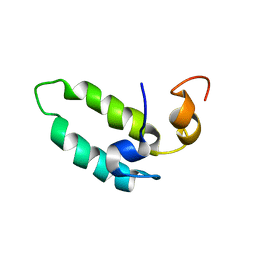 | |
1DG4
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | Descriptor: | DNAK | | Authors: | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
2BPR
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, 25 STRUCTURES | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
2V79
 
 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | Descriptor: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | Authors: | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | Deposit date: | 2007-07-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
6FFR
 
 | | DNA-RNA Hybrid Quadruplex with Flipped Tetrad | | Descriptor: | DNA/RNA (5'-R(*G)-D(P*GP*GP*AP*TP*GP*GP*GP*AP*CP*AP*CP*AP*GP*GP*GP*GP*AP*C)-R(P*G)-D(P*GP*G)-3') | | Authors: | Haase, L, Dickerhoff, J, Weisz, K. | | Deposit date: | 2018-01-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | DNA-RNA Hybrid Quadruplexes Reveal Interactions that Favor RNA Parallel Topologies.
Chemistry, 24, 2018
|
|
7U02
 
 | | Structure of the C. crescentus DriD C-domain bound to ssDNA | | Descriptor: | DNA (5'-D(P*AP*CP*G)-3'), SULFATE ION, WYL domain-containing protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
7TZV
 
 | | Structure of DriD C-domain bound to 9mer ssDNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | Authors: | Schumacher, M.A, Laub, M. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
3G8U
 
 | | DNA binding domain:GilZ 16bp complex-5 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*GP*GP*GP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*CP*CP*CP*AP*AP*TP*GP*TP*TP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
6PH5
 
 | |
4HC9
 
 | | DNA binding by GATA transcription factor-complex 3 | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*TP*TP*AP*TP*CP*TP*CP*TP*GP*AP*TP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*AP*AP*TP*CP*AP*GP*AP*GP*AP*TP*AP*AP*CP*C)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
8CEF
 
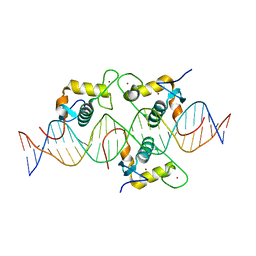 | | Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA | | Descriptor: | DNA (26-MER), Nuclear receptor DNA binding domain, ZINC ION | | Authors: | Patel, A.K.M, Shaik, T.B, McEwen, A.G, Moras, D, Klaholz, B.P, Billas, I.M.L. | | Deposit date: | 2023-02-01 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Nucleic Acids Res., 51, 2023
|
|
7PU7
 
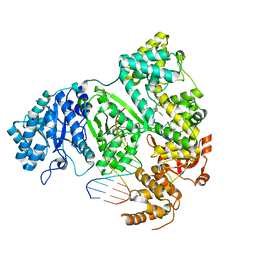 | | DNA polymerase from M. tuberculosis | | Descriptor: | DNA polymerase III subunit alpha, Template, ZINC ION, ... | | Authors: | Borsellini, A, Lamers, M.H. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DNA-Dependent Binding of Nargenicin to DnaE1 Inhibits Replication in Mycobacterium tuberculosis.
Acs Infect Dis., 8, 2022
|
|
7XV8
 
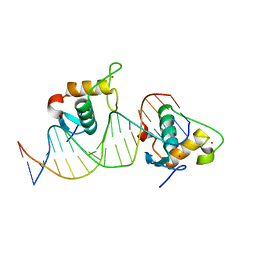 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
