6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | Descriptor: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | Authors: | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
7NHK
 
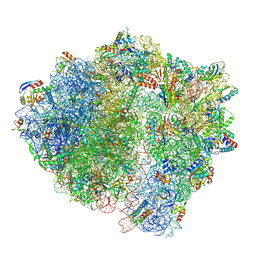 | | LsaA, an antibiotic resistance ABCF, in complex with 70S ribosome from Enterococcus faecalis | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Kasari, M, Hauryliuk, V.H, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
6FZ9
 
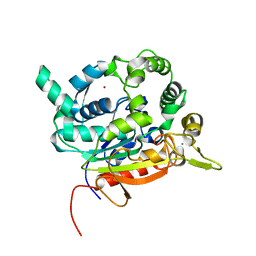 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2463 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
5Z58
 
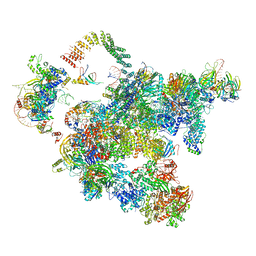 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5FCI
 
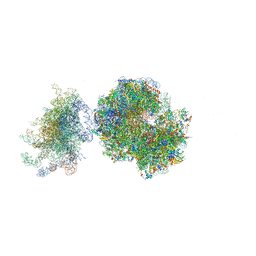 | | Structure of the vacant uL3 W255C mutant 80S yeast ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Mailliot, J, Garreau de Loubresse, N, Yusupova, G, Dinman, J.D, Yusupov, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structures of the uL3 Mutant Ribosome: Illustration of the Importance of Ribosomal Proteins for Translation Efficiency.
J.Mol.Biol., 428, 2016
|
|
5F7N
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain 17875 in complex with blood group A Lewis b pentasaccharide | | Descriptor: | Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER19, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-08 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
5FOW
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
5FKC
 
 | |
5WYJ
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
6FTK
 
 | | Gp36-MPER | | Descriptor: | Envelope protein | | Authors: | D'Ursi, A.M, Grimaldi, M. | | Deposit date: | 2018-02-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the FIV gp36 C-Terminal Heptad Repeat and Membrane-Proximal External Region.
Int J Mol Sci, 21, 2020
|
|
5WG3
 
 | | Human GRK2 in complex with Gbetagamma subunits and CCG258748 | | Descriptor: | 2-fluoro-5-[(3S,4R)-3-{[(1H-indazol-5-yl)oxy]methyl}piperidin-4-yl]-N-[(1H-pyrazol-3-yl)methyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Bouley, R, Tesmer, J.J.G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural Determinants Influencing the Potency and Selectivity of Indazole-Paroxetine Hybrid G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol. Pharmacol., 92, 2017
|
|
7M40
 
 | | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4) | | Descriptor: | Histone-binding protein RBBP4, N~3~-{4-[3-(dimethylamino)pyrrolidin-1-yl]-6,7-dimethoxyquinazolin-2-yl}-N~1~,N~1~-dimethylpropane-1,3-diamine | | Authors: | Perveen, S, Dong, A, Tempel, W, Zepeda-Velazquez, C, Abbey, M, McLeod, D, Marcellus, R, Mohammed, M, Ensan, D, Panagopoulos, D, Trush, V, Gibson, E, Brown, P.J, Arrowsmith, C.H, Schapira, M, Al-awar, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4)
To Be Published
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Dybkov, O, Kastner, B, Luehrmann, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
3NWJ
 
 | |
5WX4
 
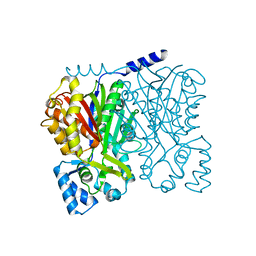 | |
7MDZ
 
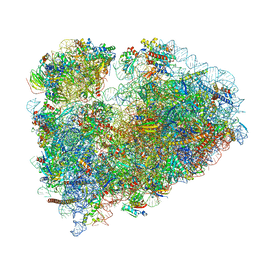 | | 80S rabbit ribosome stalled with benzamide-CHX | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S21, ... | | Authors: | Koga, Y, Hoang, E.M, Park, Y, Keszei, A.F.A, Murray, J, Shao, S, Liau, B.B. | | Deposit date: | 2021-04-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation.
J.Am.Chem.Soc., 143, 2021
|
|
5WLC
 
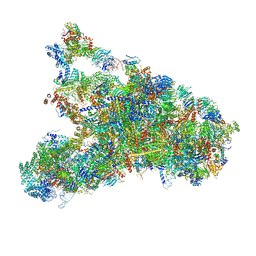 | | The complete structure of the small subunit processome | | Descriptor: | 18S pre-rRNA, 5' ETS, Bms1, ... | | Authors: | Barandun, J, Chaker-Margot, M, Hunziker, M, Klinge, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The complete structure of the small-subunit processome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3RP2
 
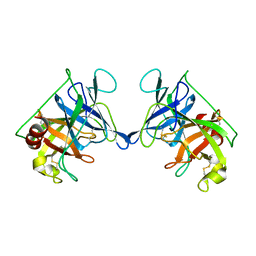 | | THE STRUCTURE OF RAT MAST CELL PROTEASE II AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | RAT MAST CELL PROTEASE II | | Authors: | Reynolds, R, Remington, S, Weaver, L, Fischer, R, Anderson, W, Ammon, H, Matthews, B. | | Deposit date: | 1984-09-10 | | Release date: | 1984-10-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of rat mast cell protease II at 1.9-A resolution.
Biochemistry, 27, 1988
|
|
6FYY
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
8CBJ
 
 | | Cryo-EM structure of Otu2-bound cytoplasmic pre-40S ribosome biogenesis complex | | Descriptor: | 20S pre-ribosomal RNA, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
7MQA
 
 | | Cryo-EM structure of the human SSU processome, state post-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
8CAS
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
6FZ7
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
5YZV
 
 | | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA | | Descriptor: | Probable serine/threonine-protein kinase PkwA | | Authors: | Li, D.Y, Shen, C, Du, Y, Qiao, F.F, Kong, T, Yuan, L.R, Zhang, D.L, Wu, X.H, Wu, Y.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA
Sci Rep, 8, 2018
|
|
