7CHN
 
 | | Crystal structure of TTK kinase domain in complex with compound 9 | | Descriptor: | 4-(cyclohexylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CLH
 
 | | Crystal structure of TTK kinase domain in complex with compound 19 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(methylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CJA
 
 | | Crystal structure of TTK kinase domain in complex with compound 28 | | Descriptor: | 4-(cyclopentylmethylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-09 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CIL
 
 | | Crystal structure of TTK kinase domain in complex with compound 7 | | Descriptor: | 4-(cyclohexylamino)-2-[(1-methylpyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
9F40
 
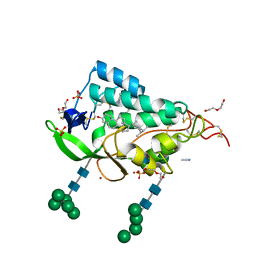 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with ergosterol bound | | Descriptor: | ACETONITRILE, DI(HYDROXYETHYL)ETHER, ERGOSTEROL, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C 1-related protein
To Be Published
|
|
9IIW
 
 | |
9F41
 
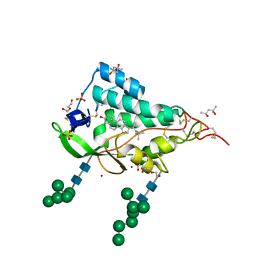 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with cholesterol bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C 1-related protein
To Be Published
|
|
9IJA
 
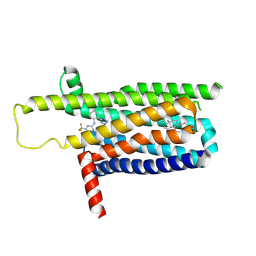 | | A local Cryo-EM structure of Bitter taste receptor TAS2R14 with Gi complex | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Taste receptor type 2 member 14 | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
6OM7
 
 | |
6ZVI
 
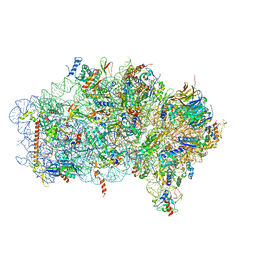 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6P5A
 
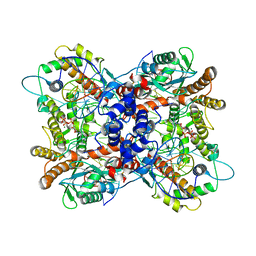 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (44-MER), DNA (5'-D(P*AP*GP*GP*TP*GP*GP*TP*CP*CP*CP*GP*TP*CP*GP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-05-30 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6P5T
 
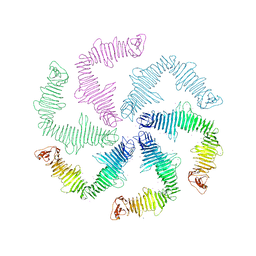 | | Surface-layer (S-layer) RsaA protein from Caulobacter crescentus bound to strontium and iodide | | Descriptor: | IODIDE ION, S-layer protein, STRONTIUM ION | | Authors: | Chan, A.C, Herrmann, J, Smit, J, Wakatsuki, S, Murphy, M.E. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bacterial surface layer protein exploits multistep crystallization for rapid self-assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y36
 
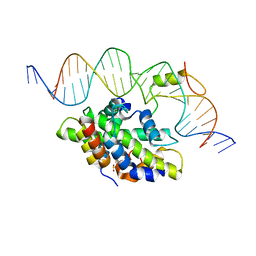 | | CCAAT-binding complex from Aspergillus fumigatus with cccA DNA | | Descriptor: | CCAAT-binding factor complex subunit HapC, CCAAT-binding factor complex subunit HapE, CCAAT-binding transcription factor subunit HAPB, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of HapE P88L -linked antifungal triazole resistance in Aspergillus fumigatus .
Life Sci Alliance, 3, 2020
|
|
8EKD
 
 | |
6Y37
 
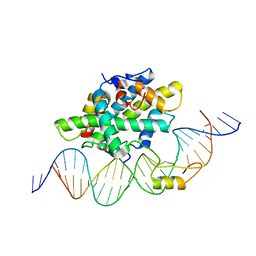 | |
6PE2
 
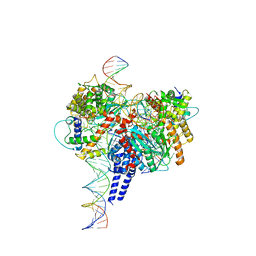 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (27-MER), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), DNA (56-MER), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Y35
 
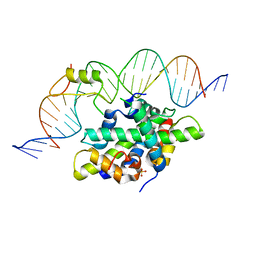 | | CCAAT-binding complex from Aspergillus fumigatus with cycA DNA | | Descriptor: | CCAAT-binding factor complex subunit HapC, CCAAT-binding factor complex subunit HapE, CCAAT-binding transcription factor subunit HAPB, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HapE P88L -linked antifungal triazole resistance in Aspergillus fumigatus .
Life Sci Alliance, 3, 2020
|
|
6PW2
 
 | |
7QI3
 
 | | Structure of Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Arylamine N-acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lowe, E.D, Kotomina, E, Karagianni, E, Boukouvala, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase is structurally, functionally and phylogenetically distinct from its N-acetyltransferase (NAT) homologues.
Febs J., 290, 2023
|
|
7ORB
 
 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7O56
 
 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element solved by Phosphorus and Sulphur SAD methods | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*AP*GP*AP*AP*AP*CP*CP*GP*AP*AP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*TP*TP*CP*GP*GP*TP*TP*TP*CP*TP*TP*TP*TP*AP*T)-3'), Interferon regulatory factor 4 | | Authors: | El Omari, K, Agnarelli, A, Duman, R, Wagner, A, Mancini, E.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-05-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorus and sulfur SAD phasing of the nucleic acid-bound DNA-binding domain of interferon regulatory factor 4.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7OXQ
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA in complex with fragment 048 at the transient P-pocket. | | Descriptor: | 2-(4-bromanylpyrazol-1-yl)-~{N}-cyclopropyl-~{N}-methyl-ethanamide, CADMIUM ION, DNA (28-MER), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
7OZ5
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA in complex with fragment 166 at the transient P-pocket. | | Descriptor: | (1~{R},2~{R})-2-phenyl-~{N}-(1,3-thiazol-2-yl)cyclopropane-1-carboxamide, CADMIUM ION, DNA (28-MER), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
7OZ2
 
 | | Crystal structure of HIV-1 reverse transcriptase with a double stranded DNA showing a transient P-pocket | | Descriptor: | CADMIUM ION, DNA (28-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Das, K. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Nat Commun, 12, 2021
|
|
8X22
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
