6DJ4
 
 | |
6DUL
 
 | |
6DRW
 
 | |
6DUW
 
 | |
4UUM
 
 | |
4USZ
 
 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
8SIL
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L, Botha, S, Poitevin, F, Sierra, R.G. | | Deposit date: | 2023-04-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SCY
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SPD
 
 | | Cytochrome P450 (CYP) 3A4 crystallized with clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Effects of Clotrimazole on X-Ray Crystal Structures of Human Cytochromes P450 3A5 and 3A4.
Drug Metab.Dispos., 51, 2023
|
|
8SYN
 
 | | Human VPS35L/VPS29/VPS26C Complex | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SYM
 
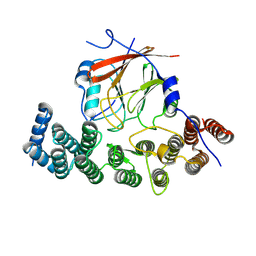 | | Human VPS29/VPS35L Complex (Locally refined map) | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SYO
 
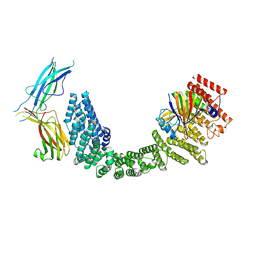 | | Human Retriever VPS35L/VPS29/VPS26C Complex (Composite Map) | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4U2T
 
 | |
4W8V
 
 | |
4UW9
 
 | |
4UWP
 
 | | Penta Zn1 coordination. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | METALLO-BETA-LACTAMASE VIM-26, ZINC ION | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
8T0B
 
 | |
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
8SD8
 
 | | Carbonic anhydrase II radiation damage RT 91-120 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4W8R
 
 | |
4W8X
 
 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | Descriptor: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
8SD7
 
 | | Carbonic anhydrase II radiation damage RT 61-90 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD6
 
 | | Carbonic anhydrase II radiation damage RT 31-60 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD9
 
 | | Carbonic anhydrase II radiation damage RT 121-150 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
