1DFL
 
 | | SCALLOP MYOSIN S1 COMPLEXED WITH MGADP:VANADATE-TRANSITION STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Houdusse, A, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 1999-11-19 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Three conformational states of scallop myosin S1.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1HLS
 
 | | NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) | | Descriptor: | INSULIN | | Authors: | Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1995-06-28 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an engineered biologically potent insulin monomer, B16 Tyr-->His, as determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1NPP
 
 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN P2(1) | | Descriptor: | ISOPROPYL ALCOHOL, Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzahn, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
7PDZ
 
 | | Structure of capping protein bound to the barbed end of a cytoplasmic actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Funk, J, Merino, F, Schacks, M, Rottner, K, Raunser, S, Bieling, P. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-01 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks.
Nat Commun, 12, 2021
|
|
1OWA
 
 | | Solution Structural Studies on Human Erythrocyte Alpha Spectrin N Terminal Tetramerization Domain | | Descriptor: | Spectrin alpha chain, erythrocyte | | Authors: | Park, S, Caffrey, M.S, Johnson, M.E, Fung, L.W. | | Deposit date: | 2003-03-28 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structural studies on human erythrocyte alpha-spectrin tetramerization site.
J.Biol.Chem., 278, 2003
|
|
7PIG
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN088 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-azanyl-2-chloranyl-3-[(Z)-piperidin-3-ylidenemethyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PID
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN060 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(3R)-morpholin-3-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIE
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN068 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(2R)-morpholin-2-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIF
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN086 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[3-[4-(aminomethyl)oxan-4-yl]phenyl]-2-azanyl-benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIH
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN093 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[3-[[(3~{R})-3-azanylpyrrolidin-1-yl]methyl]phenyl]-4~{H}-isoquinolin-1-one, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PNS
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN081 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-6-[5-[(dimethylamino)methyl]-2-fluoranyl-phenyl]-1H-indole-3-carbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
1HME
 
 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-02-10 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
7QHB
 
 | | Active state of GluA1/2 in complex with TARP gamma 8, L-glutamate and CTZ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CYCLOTHIAZIDE, ... | | Authors: | Herguedas, B, Kohegyi, B, Zhang, D, Greger, I.H. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
7QHH
 
 | | Desensitized state of GluA1/2 AMPA receptor in complex with TARP-gamma 8 (TMD-LBD) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, GLUTAMIC ACID, ... | | Authors: | Herguedas, B, Kohegyi, B, Dohrke, J.N, Watson, J.F, Zhang, D, Ho, H, Shaikh, S, Lape, R, Krieger, J.M, Greger, I.H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
1JBP
 
 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, MUSCLE/BRAIN FORM, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
7ZNL
 
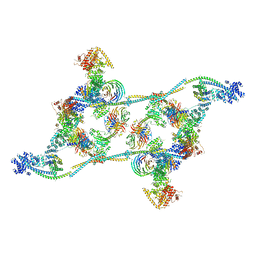 | | Structure of the human TREX core THO-UAP56 complex | | Descriptor: | Spliceosome RNA helicase DDX39B, THO complex subunit 1, THO complex subunit 2, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
1NPR
 
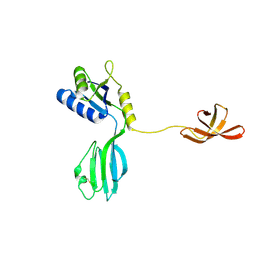 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1KKE
 
 | |
1KLX
 
 | |
1JPP
 
 | |
1JLU
 
 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
8SH5
 
 | | Crystal structure of 3'cap-independent translation enhancers (CITE) from Pea enation mosaic virus RNA 2 (PEMV2) with Fab BL3-6K170A | | Descriptor: | Fab BL3-6K170A heavy chain, Fab BL3-6K170A light chain, RNA (88-MER) | | Authors: | Lewicka, A, Roman, C, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a cap-independent translation enhancer RNA.
Nucleic Acids Res., 51, 2023
|
|
1KQQ
 
 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
1KVV
 
 | | Solution Structure Of Protein SRP19 Of The Archaeoglobus fulgidus Signal Recognition Particle, Minimized Average Structure | | Descriptor: | SRP19 | | Authors: | Pakhomova, O.N, Deep, S, Huang, Q, Zwieb, C, Hinck, A.P. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.
J.Mol.Biol., 317, 2002
|
|
1Q3T
 
 | | Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae | | Descriptor: | Cytidylate kinase | | Authors: | Yu, L, Mack, J, Hajduk, P.J, Kakavas, S.J, Saiki, A.Y, Lerner, C.G, Olejniczak, E.T. | | Deposit date: | 2003-07-31 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae
Protein Sci., 12, 2003
|
|
