2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
8RFM
 
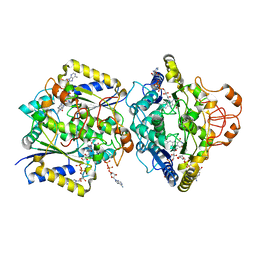 | | Human NOQ1 enzyme in complex with NADH by serial crystallography | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
8RFN
 
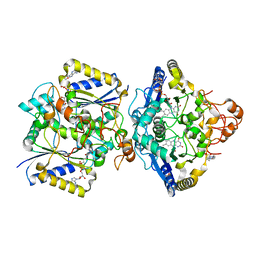 | | Human NOQ1 enzyme in its holo form by serial crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
2YHA
 
 | |
2Y8N
 
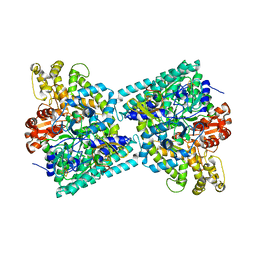 | | Crystal structure of glycyl radical enzyme | | Descriptor: | 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-08 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
2YGW
 
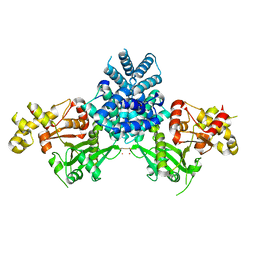 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
6ZTH
 
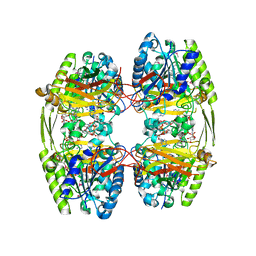 | |
2YOP
 
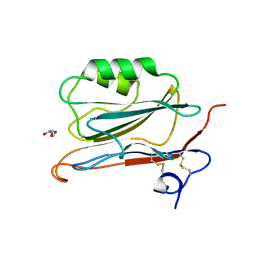 | | Long wavelength S-SAD structure of FAM3B PANDER | | Descriptor: | GLYCEROL, PROTEIN FAM3B | | Authors: | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
6ZTI
 
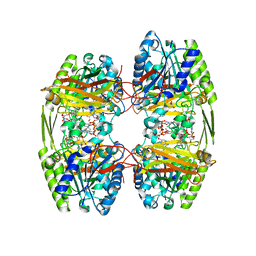 | |
3V58
 
 | |
8T1D
 
 | | Open-state cryo-EM structure of full-length human TRPV4 in complex with agonist 4a-PDD | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1F
 
 | | Cryo-EM structure of full-length human TRPV4 in complex with antagonist HC-067047 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-methyl-1-[3-(morpholin-4-yl)propyl]-5-phenyl-N-[3-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1B
 
 | | Cryo-EM structure of full-length human TRPV4 in apo state | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera, ... | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
2Y6V
 
 | |
8T1C
 
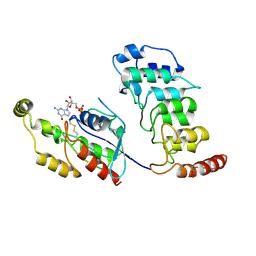 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1E
 
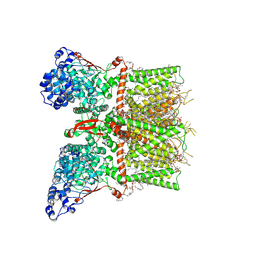 | | Closed-state cryo-EM structure of full-length human TRPV4 in the presence of 4a-PDD | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4,Enhanced green fluorescent protein, ... | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
2YAJ
 
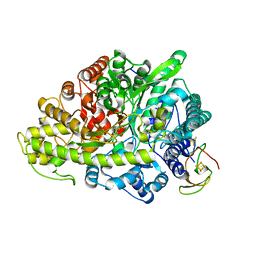 | | CRYSTAL STRUCTURE OF GLYCYL RADICAL ENZYME with bound substrate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, ... | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
7K9B
 
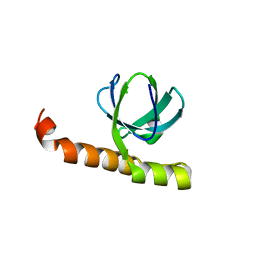 | | Crystal structure of Bacillus halodurans OapB (native) at 1.2 A | | Descriptor: | CHLORIDE ION, OLE-associated protein B | | Authors: | Yang, Y, Breaker, R.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Structure of a bacterial OapB protein with its OLE RNA target gives insights into the architecture of the OLE ribonucleoprotein complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K9C
 
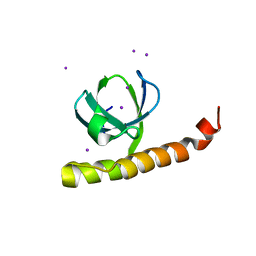 | |
7K9D
 
 | |
7K9E
 
 | |
7K7J
 
 | | EphB6 receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 6 | | Authors: | Goldgur, Y, Himanen, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the EphB6 receptor ectodomain.
Plos One, 16, 2021
|
|
7JIZ
 
 | |
7T0O
 
 | |
7T0R
 
 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | Authors: | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
