2C5B
 
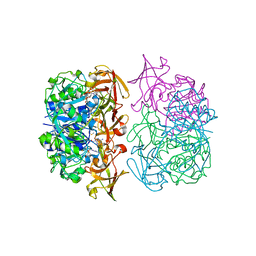 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-5'deoxy-fluoroadenosine. | | Descriptor: | 5'-FLUORO-2',5'-DIDEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.A, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity in Enzymatic Fluorination. The Fluorinase from Streptomyces Cattleya Accepts 2'-Deoxyadenosine Substrates.
Org.Biomol.Chem., 4, 2006
|
|
6X7Q
 
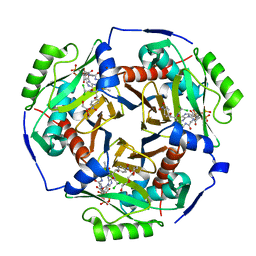 | | Chloramphenicol acetyltransferase type III in complex with chloramphenicol and acetyl-oxa(dethia)-CoA | | Descriptor: | CHLORAMPHENICOL, Chloramphenicol acetyltransferase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
7YOE
 
 | |
7YOF
 
 | |
7YOD
 
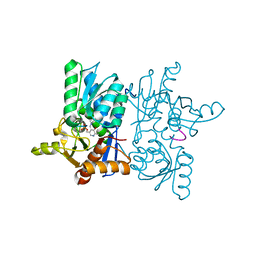 | |
7YOG
 
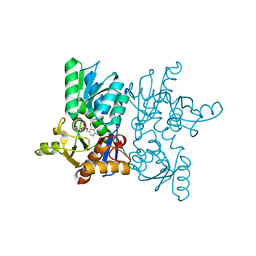 | |
7YOH
 
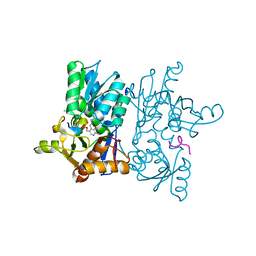 | |
7YOK
 
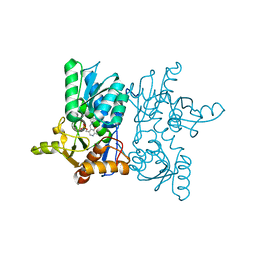 | | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A | | Descriptor: | Cysteine synthase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A
To Be Published
|
|
7YOM
 
 | | Crystal structure of tetra mutant (D67E,A68P,L98I,A301S) of O-acetylserine sulfhydrylase from Salmonella typhimurium in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.8 A | | Descriptor: | Cysteine synthase, peptide from serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of I88L single mutant of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.14 A
To Be Published
|
|
7YOI
 
 | |
7YOL
 
 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
6AIF
 
 | |
3MO7
 
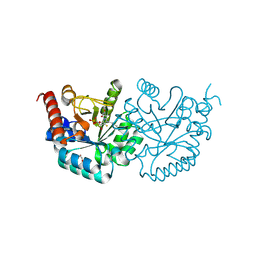 | |
4WAB
 
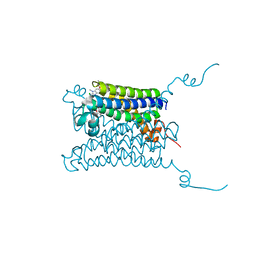 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6AE3
 
 | | Crystal structure of GSK3beta complexed with Morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, GLYCEROL, Glycogen synthase kinase-3 beta | | Authors: | Kim, K.L, Cha, J.S, Kim, J.S, Ahn, J.S, Ha, N.C, Cho, H.S. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of GSK3 beta in complex with the flavonoid, morin
Biochem. Biophys. Res. Commun., 504, 2018
|
|
8XKF
 
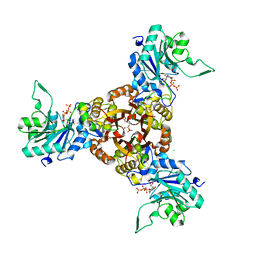 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XHU
 
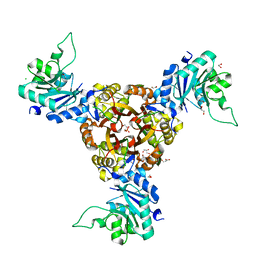 | | Crystal structure of Helicobacter pylori IspDF | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
1JEO
 
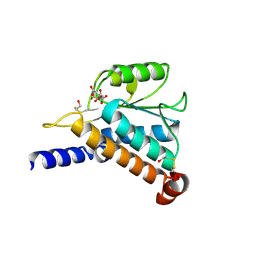 | | Crystal Structure of the Hypothetical Protein MJ1247 from Methanococcus jannaschii at 2.0 A Resolution Infers a Molecular Function of 3-Hexulose-6-Phosphate isomerase. | | Descriptor: | CITRIC ACID, HYPOTHETICAL PROTEIN MJ1247 | | Authors: | Martinez-Cruz, L.A, Dreyer, M.K, Boisvert, D.C, Yokota, H, Martinez-Chantar, M.L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-06-18 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MJ1247 protein from M. jannaschii at 2.0 A resolution infers a molecular function of 3-hexulose-6-phosphate isomerase.
Structure, 10, 2002
|
|
2C2W
 
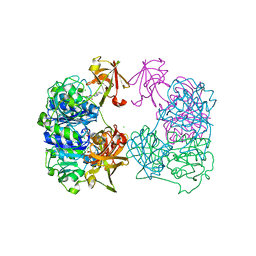 | |
8A2K
 
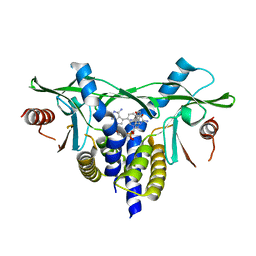 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-[(2-naphthyloxy)methyl]phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-[4-(naphthalen-1-yloxymethyl)phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
6K8C
 
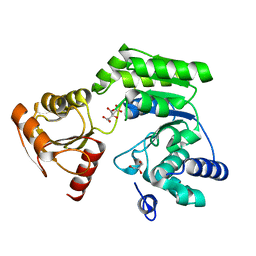 | |
1YQ7
 
 | | Human farnesyl diphosphate synthase complexed with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION, ... | | Authors: | Kavanagh, K.L, Guo, K, Von delft, F, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human farnesyl diphosphate complexed with clinical inhibitor risedronate
To be Published
|
|
7EWO
 
 | | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase | | Authors: | Rahisuddin, R, Ekka, M.K, Singh, A.K, Saini, N, Patel, M, Kumar, N, Kumaran, S. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of D67A, E68P double mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae
To Be Published
|
|
6KFU
 
 | |
7RXV
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine, Malonate (MLA) and MgF3 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, GLYCEROL, ... | | Authors: | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
