6XOQ
 
 | | DCN1 covalently bound to inhibitor 4 | | Descriptor: | (2E)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-4-(morpholin-4-yl)but-2-enamide, Lysozyme, DCN1-like protein 1 chimera | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XON
 
 | | DCN1 bound to inhibitor 9 | | Descriptor: | (2S)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-4-(dimethylamino)-2-methylbutanamide, Lysozyme DCN1-like protein 1 chimera | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOO
 
 | | DCN1 bound to DI-1859 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOM
 
 | | DCN1 bound to 8 | | Descriptor: | (2R)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-2-methyl-4-(morpholin-4-yl)butanamide, 1,2-ETHANEDIOL, Lysozyme, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOL
 
 | | DCN1 bound to DI-1548 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOP
 
 | | DCN1 bound to inhibitor 10 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-[(1S)-1-cyclohexyl-2-{[(2S)-3-(1H-imidazol-1-yl)-2-methylpropanoyl]amino}ethyl]-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XMV
 
 | | The crystal structure of Neisseria gonorrhoeae DolP (NGO1985) | | Descriptor: | CHLORIDE ION, Hemolysin,Endolysin | | Authors: | Li, J, Peterson Brown, L.E, Reed, R.W, Wierzbicki, I.H, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2020-06-30 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure and function of a new component of Bam complex in Neisseria
To Be Published
|
|
7XB5
 
 | |
6LB8
 
 | | Crystal structure of the Ca2+-free T4L-MICU1-MICU2 complex | | Descriptor: | Calcium uptake protein 2, mitochondrial, Endolysin,Calcium uptake protein 1 | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
4EPI
 
 | | The crystal structure of pesticin-T4 lysozyme hybrid stabilized by engineered disulfide bonds | | Descriptor: | Pesticin, Lysozyme Chimera, SODIUM ION, ... | | Authors: | Seddiki, N, Fairman, J.W, Noinaj, N, Lukacik, P, Barnard, T, Buchanan, S.K. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EXM
 
 | | The crystal structure of an engineered phage lysin containing the binding domain of pesticin and the killing domain of T4-lysozyme | | Descriptor: | Pesticin, Lysozyme Chimera | | Authors: | Seddiki, N, Noinaj, N, Fairman, J.W, Lukacik, P, Barnard, T.J, Buchanan, S.K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6A9E
 
 | | Crystal structure of the N-terminal domain of Atg2 | | Descriptor: | Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6A9J
 
 | | Crystal structure of the PE-bound N-terminal domain of Atg2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6AJH
 
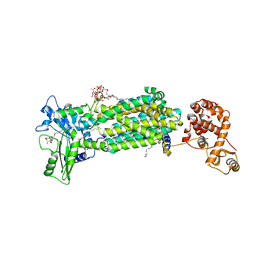 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJI
 
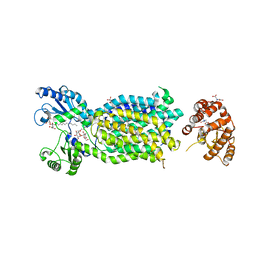 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with Rimonabant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJF
 
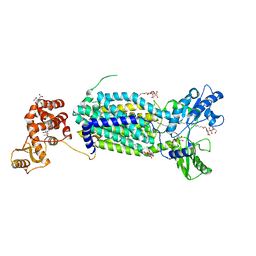 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Drug exporters of the RND superfamily-like protein,Endolysin, alpha-D-glucopyranosyl 6-O-dodecyl-alpha-D-glucopyranoside | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJG
 
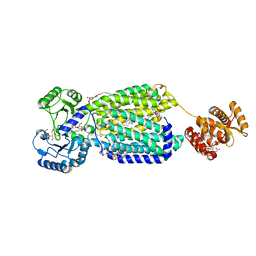 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJJ
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
5YQR
 
 | | Crystal structure of the PH-like domain of Lam6 | | Descriptor: | Endolysin/Membrane-anchored lipid-binding protein LAM6 fusion protein, NONAETHYLENE GLYCOL | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2017-11-07 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6P5W
 
 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6P5V
 
 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V88
 
 | | Structure of DCN1 bound to NAcM-COV | | Descriptor: | Lysozyme,DCN1-like protein 1, N-{2-[({1-[(2R)-pentan-2-yl]piperidin-4-yl}{[3-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]phenyl}propanamide | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
