6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
7ZD4
 
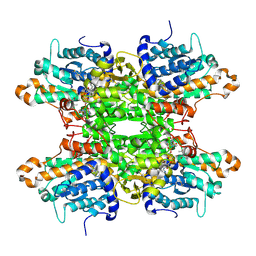 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase soaked with Cu+ ions | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD0
 
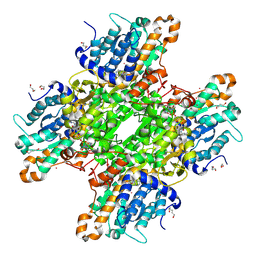 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Cd2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD1
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Hg2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD3
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Zn2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD2
 
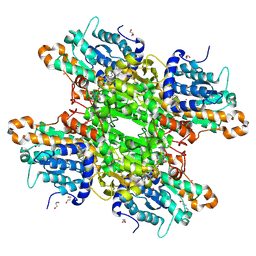 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Co2+ ions. | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
2GWD
 
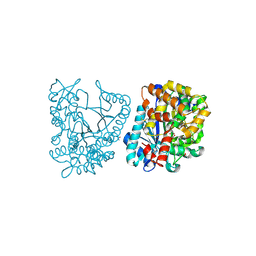 | | Crystal structure of plant glutamate cysteine ligase in complex with Mg2+ and L-glutamate | | Descriptor: | ACETATE ION, GLUTAMIC ACID, Glutamate cysteine ligase, ... | | Authors: | Hothorn, M, Wachter, A, Gromes, R, Stuwe, T, Rausch, T, Scheffzek, K. | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for the redox control of plant glutamate cysteine ligase.
J.Biol.Chem., 281, 2006
|
|
2GWC
 
 | | Crystal structure of plant glutamate cysteine ligase in complex with a transition state analogue | | Descriptor: | (2S)-2-amino-4-(S-butylsulfonimidoyl)butanoic acid, Glutamate cysteine ligase, MAGNESIUM ION | | Authors: | Hothorn, M, Wachter, A, Gromes, R, Stuwe, T, Rausch, T, Scheffzek, K. | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the redox control of plant glutamate cysteine ligase.
J.Biol.Chem., 281, 2006
|
|
7XOU
 
 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOV
 
 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
4OY3
 
 | |
3NJ4
 
 | | Fluoro-neplanocin A in Human S-Adenosylhomocysteine Hydrolase | | Descriptor: | (4S,5S)-4-(6-amino-9H-purin-9-yl)-3-fluoro-5-hydroxy-2-(hydroxymethyl)cyclopent-2-en-1-one, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jeong, L.S, Lee, K.M, Hwang, K.Y, Choi, S, Heo, Y.S. | | Deposit date: | 2010-06-17 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure and binding mode analysis of human S-adenosylhomocysteine hydrolase complexed with novel mechanism-based inhibitors, haloneplanocin A analogues.
J.Med.Chem., 54, 2011
|
|
3LGS
 
 | |
5B7N
 
 | |
5BP9
 
 | | Crystal structure of SAM-dependent methyltransferase from Bacteroides fragilis in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Putative methyltransferase protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Cymborowski, M.T, Mason, D.V, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase fromBacteroides fragilis in complex with S-Adenosyl-L-homocysteine
to be published
|
|
1X1D
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn-bacteriopheophorbide d | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-04 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1B
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
4F2P
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with diEtglycol-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[2-(2-hydroxyethoxy)ethyl]sulfanyl}methyl)pyrrolidin-3- ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with diEtglycol-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
5BP7
 
 | | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kutner, J, Shabalin, I.G, Mason, D.V, Handing, K.B, Gasiorowska, O.A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine
to be published
|
|
1X1C
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn2+ | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
4F3C
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
8HFQ
 
 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | Authors: | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4F1W
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with Adenine | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with Adenine
Structure, 21, 2013
|
|
