8QB3
 
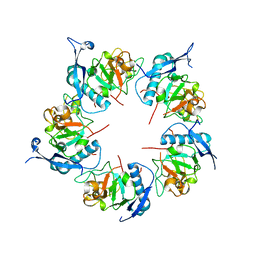 | | ADDobody zinc containing condition | | Descriptor: | ADDobody, ZINC ION | | Authors: | Buzas, D, Toelzer, C, Berger, I, Schaffitzel, C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QBX
 
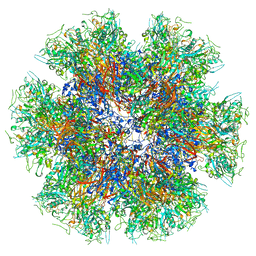 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8Q0N
 
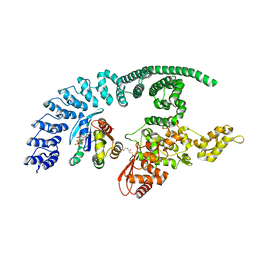 | | HACE1 in complex with RAC1 Q61L | | Descriptor: | E3 ubiquitin-protein ligase HACE1, GUANOSINE-5'-TRIPHOSPHATE, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Wolter, M, Duering, J, Dienemann, C, Lorenz, S. | | Deposit date: | 2023-07-28 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QCF
 
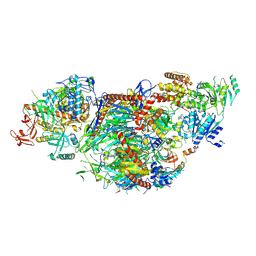 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8PWL
 
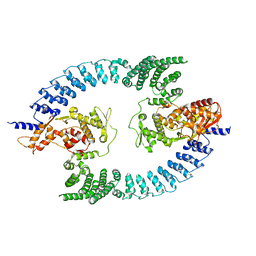 | |
5AGE
 
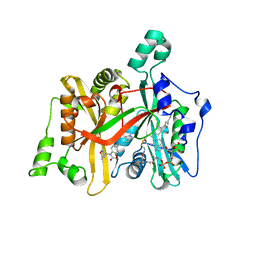 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINONE LIGAND | | Descriptor: | 4-[(5-methyl-1,2-oxazol-3-yl)methyl]-7-[4-(1-methylpiperidin-4-yl)butyl]-2H-1,4-benzoxazin-3(4H)-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q5H
 
 | | Human KMN network (outer kinetochore) | | Descriptor: | Kinetochore protein Spc24, Kinetochore protein Spc25, Kinetochore scaffold 1, ... | | Authors: | Raisch, T, Polley, S, Vetter, I, Musacchio, A, Raunser, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the human KMN complex and implications for regulation of its assembly.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5AIU
 
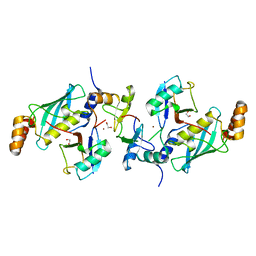 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
5QBX
 
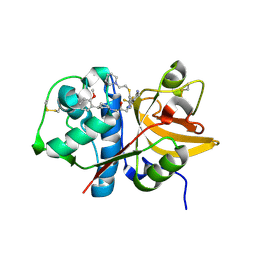 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | (2S)-1-[4-(2-methoxyphenyl)piperidin-1-yl]-3-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propan-2-ol, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCE
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{2-chloro-5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
3IXF
 
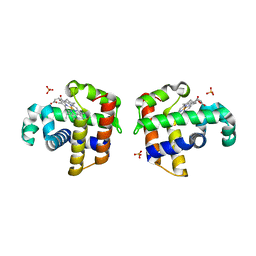 | | Crystal Structure of Dehaloperoxidase B at 1.58 and Structural Characterization of the AB Dimer from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, D'Antonio, J, Thompson, M.K, Franzen, S, Ghiladi, R.A. | | Deposit date: | 2009-09-03 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of dehaloperoxidase B at 1.58 A resolution and structural characterization of the AB dimer from Amphitrite ornata.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5QCA
 
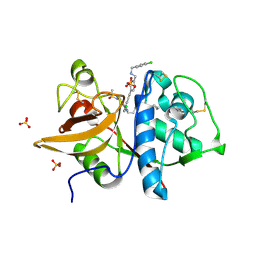 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{4-[(2-chloro-5-{1-[3-(4-cyclopropylpiperazin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}-N-[(4-chlorophenyl)methyl]methanamine, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
8QCS
 
 | |
8Q7E
 
 | |
8QCT
 
 | |
8QCX
 
 | |
8QAF
 
 | |
5B07
 
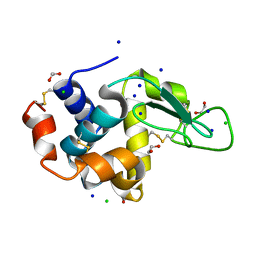 | | Lysozyme (denatured by DCl and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
8PVX
 
 | |
5QBU
 
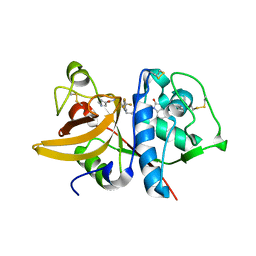 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[1-(3-{5-(1H-imidazole-5-carbonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl)piperidin-4-yl]-3-methyl-1,3-dihydro-2H-benzimidazol-2-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
3IQC
 
 | |
4ZEE
 
 | |
4ZEQ
 
 | | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide, Northeast Structural Genomics Consortium Target HX9247 | | Descriptor: | BH3-interacting domain death agonist, Bcl-2-related protein A1 | | Authors: | Guan, R, Xiao, R, Mao, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide
To Be Published
|
|
3FRS
 
 | | Structure of human IST1(NTD) (residues 1-189)(p43212) | | Descriptor: | GLYCEROL, Uncharacterized protein KIAA0174 | | Authors: | Schubert, H.L, Hill, C.P, Bajorek, M, Sundquist, W.I. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for ESCRT-III protein autoinhibition.
Nat.Struct.Mol.Biol., 16, 2009
|
|
