4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | Descriptor: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
4UD0
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UFD
 
 | | Thrombin in complex with 4-(((1-((2S)-1-((2R)-2-(benzylsulfonylamino)- 3-phenyl-propanoyl)pyrrolidin-2-yl)-1-oxo-ethyl)amino)methyl) benzamidine | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2R)-3-phenyl-2-[(phenylmethyl)sulfonylamino]propanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
4UFG
 
 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-((1S)-2-((4- carbamimidoylphenyl)methylamino)-1-methyl-2-oxo-ethyl)-N-methyl-3- phenyl-propanamide ethane | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-2-[methyl-[(2R)-3-phenyl-2-[(phenylmethyl)sulfonylamino]propanoyl]amino]butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-2, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
4UCL
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCY
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UGO
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N-(4-(2-(ethyl(3-(((E)-imino(thiophen-2-yl)methyl)amino)benzyl)amino) ethyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4KSC
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6QNH
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 0ms timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
4UFE
 
 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-(2-((4- carbamimidoylphenyl)methylamino)-2-oxo-butyl)-3-phenyl-propanamide | | Descriptor: | (2R)-N-[(2S)-1-[(4-carbamimidoylphenyl)methylamino]-1-oxidanylidene-propan-2-yl]-3-phenyl-2-[(phenylmethyl)sulfonylamino]propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-2, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
4K14
 
 | |
4K2L
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92Q at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cavity hydration in proteins
To be Published
|
|
6QOE
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 8 (3-[(2-thienylthio)methyl]benzoic acid) | | Descriptor: | 3-(thiophen-2-ylsulfanylmethyl)benzoic acid, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
4KJJ
 
 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
6DRV
 
 | |
4UCJ
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4KSB
 
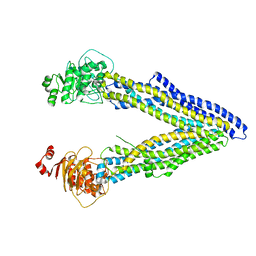 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4UCI
 
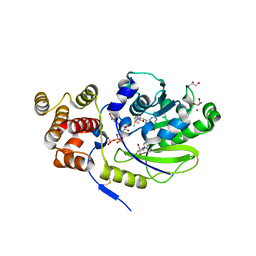 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | ADENOSINE, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-10-14 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4K48
 
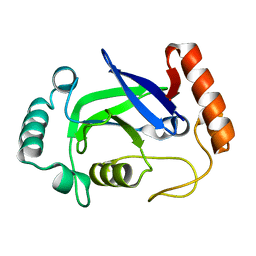 | | Structure of the Streptococcus pneumoniae leucyl-tRNA synthetase editing domain | | Descriptor: | Leucine--tRNA ligase | | Authors: | Hu, Q.H, Liu, R.J, Fang, Z.P, Zhang, J, Ding, Y.Y, Tan, M, Wang, M, Pan, W, Zhou, H.C, Wang, E.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase
Sci Rep, 3, 2013
|
|
4KD1
 
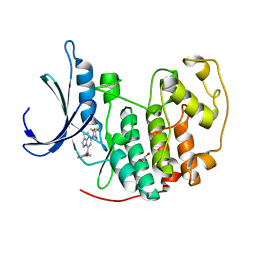 | | CDK2 in complex with Dinaciclib | | Descriptor: | 1,2-ETHANEDIOL, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Cyclin-dependent kinase 2 | | Authors: | Martin, M.P, Schonbrunn, E. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains.
Acs Chem.Biol., 8, 2013
|
|
4UJ4
 
 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab-3A-interacting protein, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of Rab11-FIP3-Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11.
Nat. Struct. Mol. Biol., 22, 2015
|
|
4UJ3
 
 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6QPP
 
 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
