2W5F
 
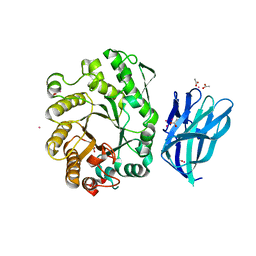 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2008-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
3DEK
 
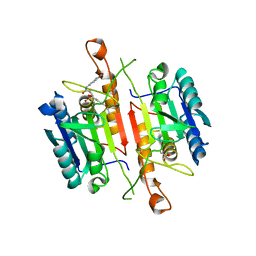 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, N-[3-(2-fluoroethoxy)phenyl]-N'-(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-6-yl)butanediamide | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
2F6Y
 
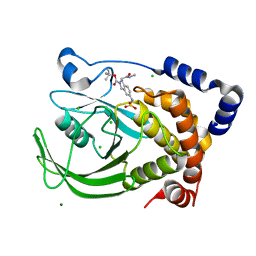 | | Protein tyrosine phosphatase 1B with sulfamic acid inhibitors | | Descriptor: | 3(R)-METHYLCARBAMOYL-7-SULFOAMINO-3,4-DIHYDRO-1H-ISOQUINOLINE-2-CARBOXYLIC ACID TERT-BUTYL ESTER, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Klopfenstein, S.R. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 1,2,3,4-Tetrahydroisoquinolinyl sulfamic acids as phosphatase PTP1B inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3DNL
 
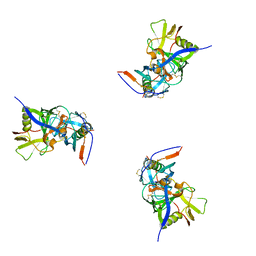 | | Molecular structure for the HIV-1 gp120 trimer in the b12-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
1Z2O
 
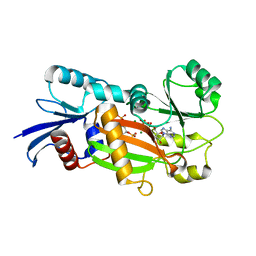 | | Inositol 1,3,4-trisphosphate 5/6-Kinase in complex with mg2+/ADP/Ins(1,3,4,6)P4 | | Descriptor: | (1S,3R,4R,6S)-1,3,4,6-TETRAPKISPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
3T2F
 
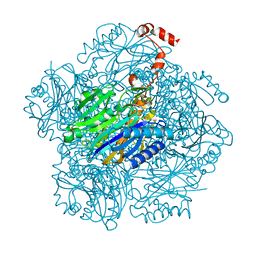 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, soaked with EDTA and DHAP | | Descriptor: | Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3OTS
 
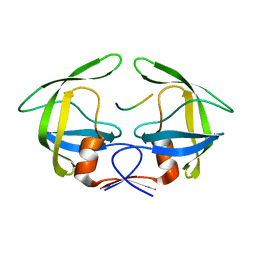 | | MDR769 HIV-1 protease complexed with MA/CA hepta-peptide | | Descriptor: | MA/CA substrate peptide, Multi-drug resistant HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
2FYD
 
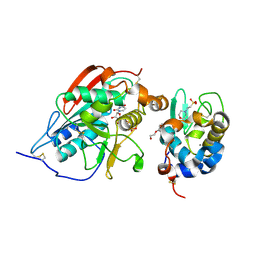 | | catalytic domain of bovine beta 1, 4-galactosyltransferase in complex with alpha-lactalbumin, glucose, Mn, and UDP-N-acetylgalactosamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-galactopyranose, Alpha-lactalbumin, ... | | Authors: | Ramakrishnan, B, Ramasamy, V, Qasba, P.K. | | Deposit date: | 2006-02-07 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Snapshots of beta-1,4-Galactosyltransferase-I Along the Kinetic Pathway.
J.Mol.Biol., 357, 2006
|
|
2L19
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Arsenate reductase | | Authors: | Yu, C, Xia, B, Jin, C. | | Deposit date: | 2010-07-26 | | Release date: | 2011-04-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803
Biomol.Nmr Assign., 5, 2011
|
|
3OTY
 
 | | MDR769 HIV-1 protease complexed with RT/RH hepta-peptide | | Descriptor: | MDR HIV-1 protease, RT/RH substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUA
 
 | | MDR769 HIV-1 protease complexed with p1/p6 hepta-peptide | | Descriptor: | HIV-1 protease, p1/p6 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
2R3E
 
 | |
1JJE
 
 | |
3UJI
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2558 in complex with MN peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein gp160, ... | | Authors: | Kong, X.P. | | Deposit date: | 2011-11-07 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Anti-V3 HIV-1 Monoclonal Antibodies Encoded by the VH5-51/VL Lambda Genes Define a Conserved Antigenic Structure.
Plos One, 6, 2011
|
|
3QBH
 
 | | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors | | Descriptor: | (4S)-4-(2-hydroxy-5-{[(3S,4S,5R)-4-hydroxy-1,1-dioxido-5-{[3-(propan-2-yl)benzyl]amino}tetrahydro-2H-thiopyran-3-yl]methyl}benzyl)-3-propyl-1,3-oxazolidin-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2D1O
 
 | | Stromelysin-1 (MMP-3) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, SM-25453, Stromelysin-1, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
1BIU
 
 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4QK4
 
 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to pip2 at 2.8 a resolution | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-05 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3T2E
 
 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, F6P-bound form | | Descriptor: | FRUCTOSE -6-PHOSPHATE, Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
1BV9
 
 | | HIV-1 PROTEASE (I84V) COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
3OI7
 
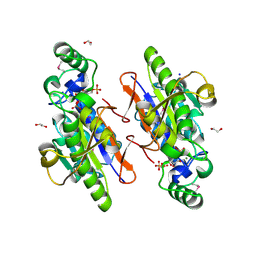 | | Structure of the structure of the H13A mutant of Ykr043C in complex with sedoheptulose-1,7-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,7-di-O-phosphono-beta-D-altro-hept-2-ulofuranose, GLYCEROL, ... | | Authors: | Singer, A.U, Xu, X, Dong, A, Cui, H, Clasquin, M.F, Caudy, A.A, Edwards, A.M, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Riboneogenesis in yeast.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QGE
 
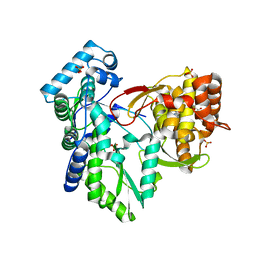 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QIO
 
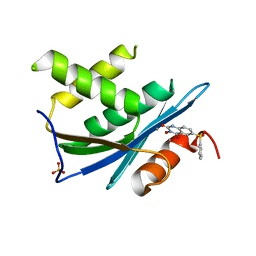 | | Crystal Structure of HIV-1 RNase H with engineered E. coli loop and N-hydroxy quinazolinedione inhibitor | | Descriptor: | 3-hydroxy-6-(phenylsulfonyl)quinazoline-2,4(1H,3H)-dione, Gag-Pol polyprotein,Ribonuclease HI,Gag-Pol polyprotein, MANGANESE (II) ION, ... | | Authors: | Lansdon, E.B, Liu, Q. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4011 Å) | | Cite: | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
2JIW
 
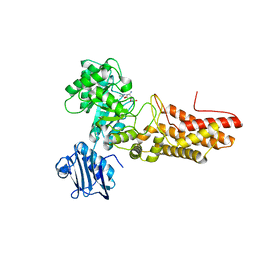 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine | | Descriptor: | N-[(1S,2R,5R,6R)-2-AMINO-5,6-DIHYDROXY-4-(HYDROXYMETHYL)CYCLOHEX-3-EN-1-YL]ACETAMIDE, O-GLCNACASE BT_4395 | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A 1-Acetamido Derivative of 6-Epi-Valienamine: An Inhibitor of a Diverse Group of Beta-N-Acetylglucosaminidases.
Org.Biomol.Chem., 5, 2007
|
|
1XDD
 
 | | X-ray structure of LFA-1 I-domain in complex with LFA703 at 2.2A resolution | | Descriptor: | 8-[2-((2S)-4-HYDROXY-1-{[5-(HYDROXYMETHYL)-6-METHOXY-2-NAPHTHYL]METHYL}-6-OXOPIPERIDIN-2-YL)ETHYL]-3,7-DIMETHYL-1,2,3,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL 2-METHYLBUTANOATE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Weitz-Schmidt, G, Welzenbach, K, Dawson, J, Kallen, J. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved lymphocyte function-associated antigen-1 (LFA-1) inhibition by statin derivatives: molecular basis determined by x-ray analysis and monitoring of LFA-1 conformational changes in vitro and ex vivo
J.Biol.Chem., 279, 2004
|
|
