2PKN
 
 | | Crystal structure of M tuberculosis Adenosine Kinase complexed with AMP-PCP (non-hydrolyzable ATP analog) | | Descriptor: | Adenosine kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
1PBP
 
 | | FINE TUNING OF THE SPECIFICITY OF THE PERIPLASMIC PHOSPHATE TRANSPORT RECEPTOR: SITE-DIRECTED MUTAGENESIS, LIGAND BINDING, AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Wang, Z, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1994-07-20 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies.
J.Biol.Chem., 269, 1994
|
|
2PKF
 
 | | Crystal structure of M tuberculosis Adenosine Kinase (apo) | | Descriptor: | Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
1QUI
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH BROMINE AND PHOSPHATE | | Descriptor: | BROMIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1IW6
 
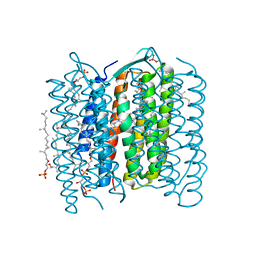 | | Crystal Structure of the Ground State of Bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Kouyama, T, Okumura, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-22 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary
Photoreaction of Bacteriorhodopsin.
J.Mol.Biol., 324, 2002
|
|
1IXF
 
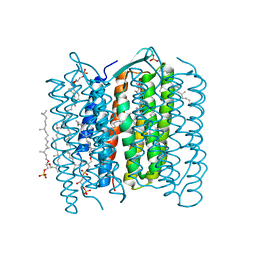 | | Crystal Structure of the K intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Matsui, Y, Sakai, K, Murakami, M, Shiro, Y, Adachi, S, Okumura, H, Kouyama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-20 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin
J.MOL.BIOL., 324, 2002
|
|
1SJQ
 
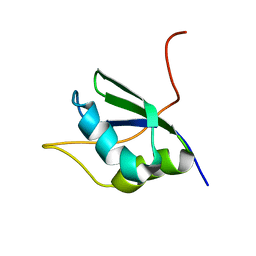 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1II1
 
 | |
1IJ6
 
 | | CA2+-BOUND STRUCTURE OF MULTIDOMAIN EF-HAND PROTEIN, CBP40, FROM TRUE SLIME MOLD | | Descriptor: | CALCIUM ION, PLASMODIAL SPECIFIC LAV1-2 PROTEIN | | Authors: | Iwasaki, W, Sasaki, H, Nakamura, A, Kohama, K, Tanokura, M. | | Deposit date: | 2001-04-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Metal-Free and Ca(2+)-Bound Structures of a Multidomain EF-Hand Protein, CBP40, from the Lower Eukaryote Physarum polycephalum
Structure, 11, 2003
|
|
1WWQ
 
 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
1T5H
 
 | |
1IJ5
 
 | | METAL-FREE STRUCTURE OF MULTIDOMAIN EF-HAND PROTEIN, CBP40, FROM TRUE SLIME MOLD | | Descriptor: | PLASMODIAL SPECIFIC LAV1-2 PROTEIN | | Authors: | Iwasaki, W, Sasaki, H, Nakamura, A, Kohama, K, Tanokura, M. | | Deposit date: | 2001-04-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Metal-Free and Ca(2+)-Bound Structures of a Multidomain EF-Hand Protein, CBP40, from the Lower Eukaryote Physarum polycephalum
Structure, 11, 2003
|
|
1O86
 
 | | Crystal Structure of Human Angiotensin Converting Enzyme in complex with lisinopril. | | Descriptor: | ANGIOTENSIN CONVERTING ENZYME, CHLORIDE ION, GLYCINE, ... | | Authors: | Natesh, R, Schwager, S.L.U, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2002-11-25 | | Release date: | 2003-02-07 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Angiotensin-Converting Enzyme-Lisinopril Complex
Nature, 421, 2003
|
|
1QUL
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY THR COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUJ
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUK
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY ASN COMPLEX WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1TAN
 
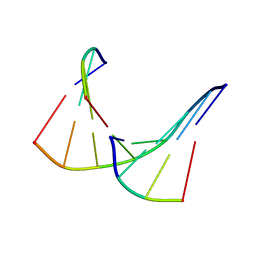 | | TANDEM DNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(P*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*AP*GP*CP*TP*G)-3') | | Authors: | Denisov, A, Sandstrom, A, Maltseva, T, Pyshnyi, D, Ivanova, E, Zarytova, V, Chattopadhyaya, J. | | Deposit date: | 1997-06-17 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of estrone (Es)-tethered tandem DNA duplex: [d(5'pCAGCp3')-Es] + [Es-d(5'pTCCA3')]: d(5'pTGGAGCTG3').
J.Biomol.Struct.Dyn., 15, 1997
|
|
1TBZ
 
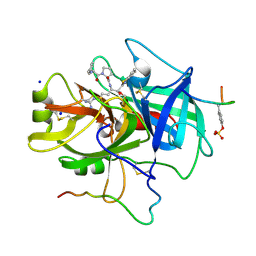 | | HUMAN THROMBIN WITH ACTIVE SITE N-METHYL-D PHENYLALANYL-N-[5-(AMINOIMINOMETHYL)AMINO]-1-{{BENZOTHIAZOLYL)CARBONYL] BUTYL]-L-PROLINAMIDE TRIFLUROACETATE AND EXOSITE-HIRUGEN | | Descriptor: | ALPHA-THROMBIN, D-phenylalanyl-N-{(1S)-1-[(S)-1,3-benzothiazol-2-yl(hydroxy)methyl]-4-carbamimidamidobutyl}-L-prolinamide, HIRUGEN, ... | | Authors: | Matthews, J.H, Krishnan, R, Costanzo, M.J, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of thrombin with thiazole-containing inhibitors: probes of the S1' binding site.
Biophys.J., 71, 1996
|
|
1VZX
 
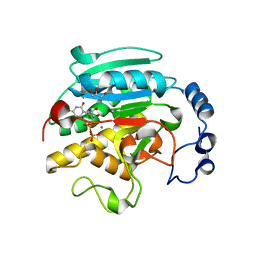 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-28 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1JN4
 
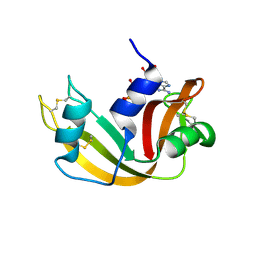 | | The Crystal Structure of Ribonuclease A in complex with 2'-deoxyuridine 3'-pyrophosphate (P'-5') adenosine | | Descriptor: | ADENOSINE-5'-[TRIHYDROGEN DIPHOSPHATE] P'-3'-ESTER WITH 2'-DEOXYURIDINE, Pancreatic Ribonuclease A | | Authors: | Jardine, A.M, Leonidas, D.D, Jenkins, J.L, Park, C, Raines, R.T, Acharya, K.R, Shapiro, R. | | Deposit date: | 2001-07-23 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cleavage of 3',5'-Pyrophosphate-Linked Dinucleotides by Ribonuclease A and Angiogenin
Biochemistry, 40, 2001
|
|
1O8A
 
 | | Crystal Structure of Human Angiotensin Converting Enzyme (Native). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Natesh, R, Schwager, S.L.U, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2002-11-26 | | Release date: | 2003-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human angiotensin-converting enzyme-lisinopril complex.
Nature, 421, 2003
|
|
1GWE
 
 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2N5W
 
 | | The NMR solution structure of octyl-tridecaptin A1 in DPC micelles | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Acedo, J.Z, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N24
 
 | |
1OWC
 
 | | Three Dimensional Structure Analysis Of The R109L Variant of the Type II Citrate Synthase From E. Coli | | Descriptor: | Citrate synthase, SULFATE ION | | Authors: | Stokell, D.J, Donald, L.J, Maurus, R, Nguyen, N.T, Sadler, G, Choudhary, K, Hultin, P.G, Brayer, G.D, Duckworth, H.W. | | Deposit date: | 2003-03-28 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the roles of key residues in the unique regulatory NADH binding site of type II citrate synthase of Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
