1NCR
 
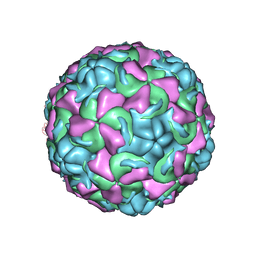 | | The structure of Rhinovirus 16 when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, MYRISTIC ACID, ZINC ION, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-05 | | Release date: | 2003-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
1NCQ
 
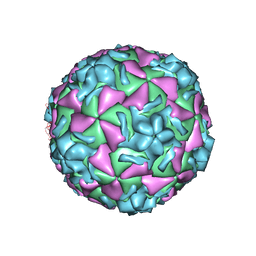 | | The structure of HRV14 when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, COAT PROTEIN VP1, COAT PROTEIN VP2, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-05 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
7WUU
 
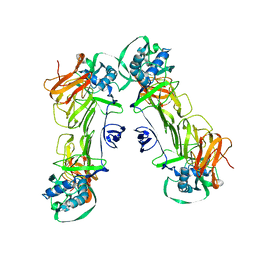 | | CryoEM structure of loose sNS1 tetramer | | Descriptor: | Core protein | | Authors: | Shu, B, Lok, S.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Nat Commun, 13, 2022
|
|
4WV4
 
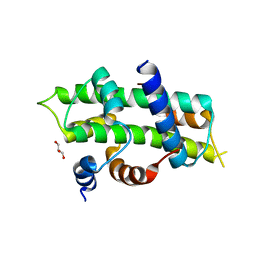 | | Heterodimer of TAF8/TAF10 | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Trowitzsch, S. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules.
Nat Commun, 6, 2015
|
|
7WUV
 
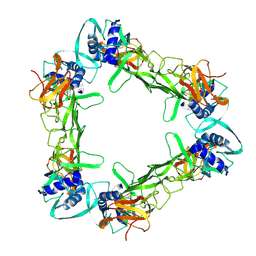 | | CryoEM structure of sNS1 hexamer | | Descriptor: | Core protein | | Authors: | Shu, B, Ooi, J.S.G, Lok, S.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Nat Commun, 13, 2022
|
|
7WUT
 
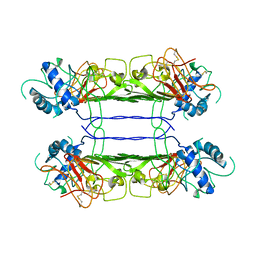 | |
2H24
 
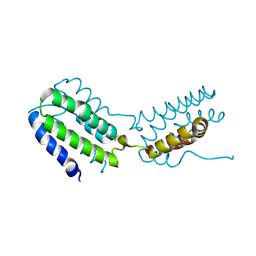 | | Crystal structure of human IL-10 | | Descriptor: | Interleukin-10 | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2006-05-18 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex.
J.Biol.Chem., 281, 2006
|
|
3MU6
 
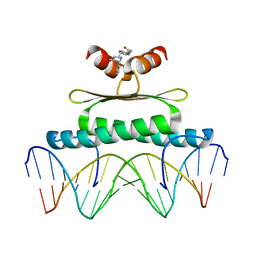 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
4MG3
 
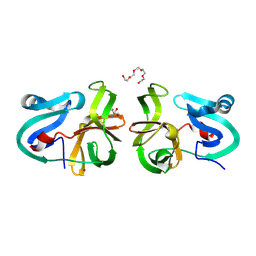 | | Crystal Structural Analysis of 2A Protease from Coxsackievirus A16 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protease 2A, ... | | Authors: | Sun, Y, Wang, X, Dang, M, Yuan, S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | An open conformation determined by a structural switch for 2A protease from coxsackievirus A16.
Protein Cell, 4, 2013
|
|
2CXV
 
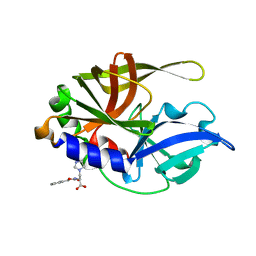 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
2B0F
 
 | |
8G0Z
 
 | |
5HRK
 
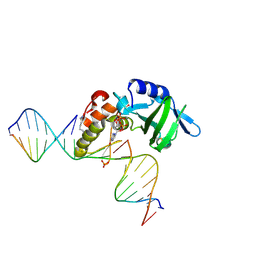 | |
5HRF
 
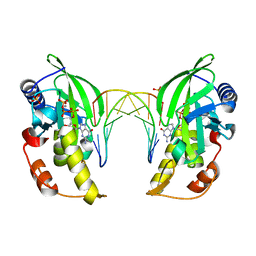 | |
5HRL
 
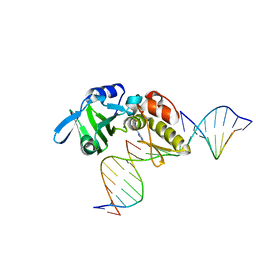 | |
1ZQ3
 
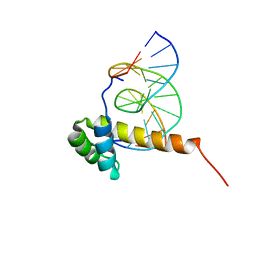 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
5FP5
 
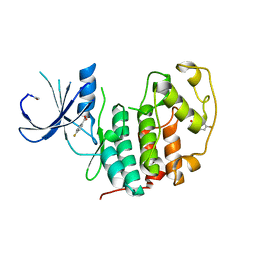 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 4- fluorobenzoic acid (AT222) in an alternate binding site. | | Descriptor: | 4-fluorobenzoic acid, ACETYL GROUP, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3K44
 
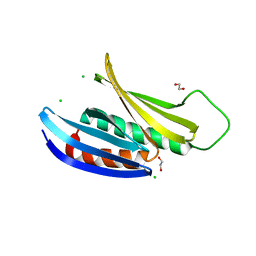 | | Crystal Structure of Drosophila melanogaster Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Purine-rich binding protein-alpha, ... | | Authors: | Graebsch, A, Roche, S, Niessing, D. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5HRH
 
 | |
3QYZ
 
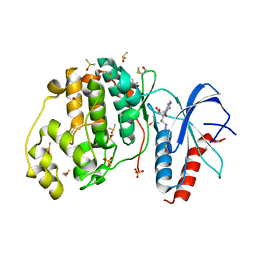 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3QYW
 
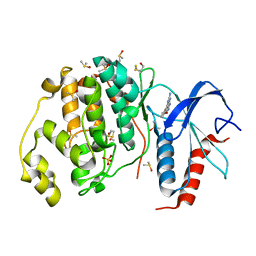 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 6-(3-bromophenyl)-7H-purin-2-amine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1AWC
 
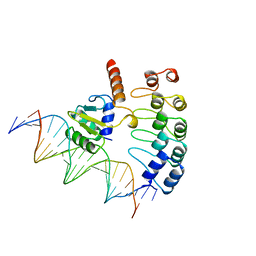 | | MOUSE GABP ALPHA/BETA DOMAIN BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*(BRU)P*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*AP*(CBR)P*AP*CP*(CBR)P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*GP*(BRU)P*GP*(BRU)P*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*AP*T)-3'), PROTEIN (GA BINDING PROTEIN ALPHA), ... | | Authors: | Batchelor, A.H, Wolberger, C. | | Deposit date: | 1997-10-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA.
Science, 279, 1998
|
|
2H9H
 
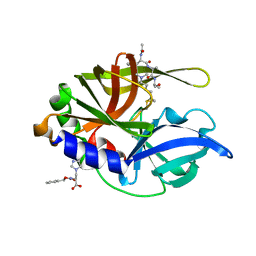 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
7O3D
 
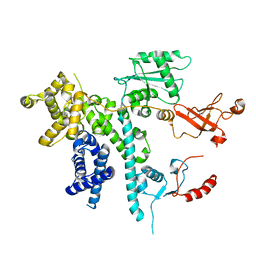 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7O6B
 
 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
