7MXG
 
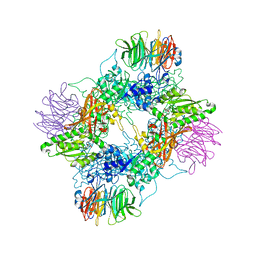 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7NA8
 
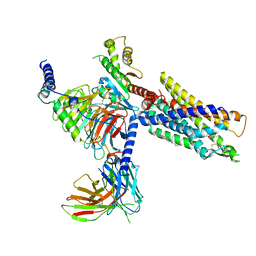 | | Structures of human ghrelin receptor-Gi complexes with ghrelin and a synthetic agonist | | Descriptor: | 1-(methanesulfonyl)-1'-(2-methyl-L-alanyl-O-benzyl-D-seryl)-1,2-dihydrospiro[indole-3,4'-piperidine], Antibody fragment, CHOLESTEROL, ... | | Authors: | Liu, H, Sun, D, Sun, J, Zhang, C. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human ghrelin receptor signaling by ghrelin and the synthetic agonist ibutamoren
Nat Commun, 12, 2021
|
|
7NA7
 
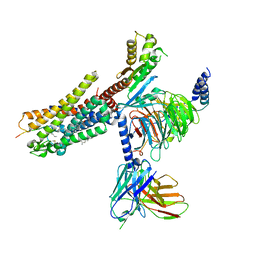 | | Structures of human ghrelin receptor-Gi complexes with ghrelin and a synthetic agonist | | Descriptor: | Antibody fragment, CHOLESTEROL, Ghrelin-27, ... | | Authors: | Liu, H, Sun, D, Sun, J, Zhang, C. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human ghrelin receptor signaling by ghrelin and the synthetic agonist ibutamoren
Nat Commun, 12, 2021
|
|
5I2T
 
 | |
7N40
 
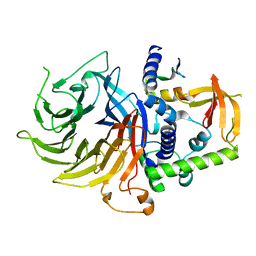 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
6EM3
 
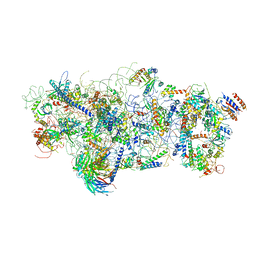 | | State A architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EG8
 
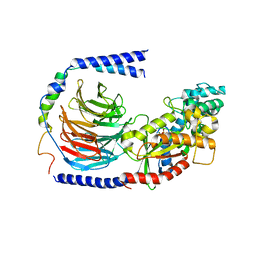 | | Structure of the GDP-bound Gs heterotrimer | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hilger, D, Liu, X, Aschauer, P, Kobilka, B.K. | | Deposit date: | 2018-08-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
5IGO
 
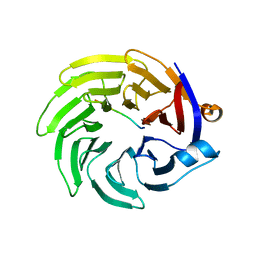 | |
7N8B
 
 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7N61
 
 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5IJ7
 
 | | Structure of Hs/AcPRC2 in complex with a pyridone inhibitor | | Descriptor: | 5,8-dichloro-2-[(4-ethyl-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-({1-[(2R)-2-hydroxypropanoyl]piperidin-4-yl}oxy)-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of Zeste Homolog 2 (EZH2),Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Deng, Y.-L, Liu, W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Nat Commun, 7, 2016
|
|
6FAI
 
 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
5I72
 
 | | Crystal structure of the oligomeric form of the Lassa virus matrix protein Z | | Descriptor: | RING finger protein Z, ZINC ION | | Authors: | Hastie, K, Zandonatti, M, Liu, T, Li, S, Woods Jr, V, Saphire, E.O. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z.
J.Virol., 90, 2016
|
|
4LYZ
 
 | |
7NRD
 
 | | Structure of the yeast Gcn1 bound to a colliding stalled 80S ribosome with MBF1, A/P-tRNA and P/E-tRNA | | Descriptor: | 25S rRNA (3184-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5IGQ
 
 | |
6FF7
 
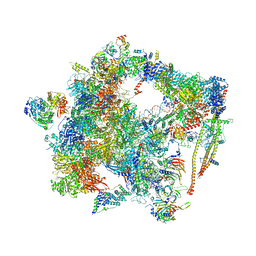 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
6F0X
 
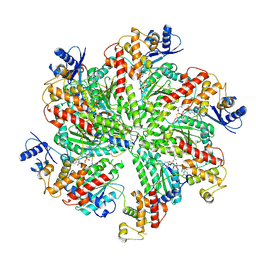 | | Cryo-EM structure of TRIP13 in complex with ATP gamma S, p31comet, C-Mad2 and Cdc20 | | Descriptor: | Cell division cycle protein 20 homolog, MAD2L1-binding protein, Mitotic spindle assembly checkpoint protein MAD2A, ... | | Authors: | Alfieri, C, Chang, L, Barford, D. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13.
Nature, 559, 2018
|
|
5IPB
 
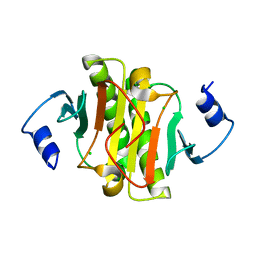 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1 | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
7OHQ
 
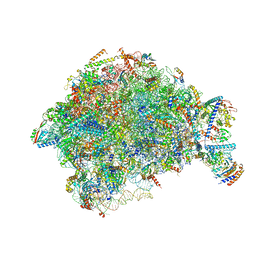 | |
7OHS
 
 | |
7OH3
 
 | |
7OHW
 
 | |
7OHP
 
 | |
7OHX
 
 | |
