1AIR
 
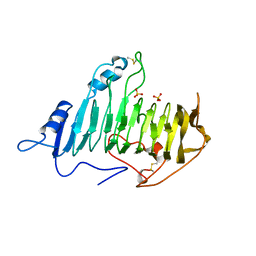 | | PECTATE LYASE C FROM ERWINIA CHRYSANTHEMI (EC16) TO A RESOLUTION OF 2.2 ANGSTROMS WITH 128 WATERS | | Descriptor: | PECTATE LYASE C, SULFATE ION | | Authors: | Lietzke, S.E, Scavetta, R.D, Yoder, M.D, Jurnak, F.A. | | Deposit date: | 1997-04-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Refined Three-Dimensional Structure of Pectate Lyase E from Erwinia chrysanthemi at 2.2 A Resolution.
Plant Physiol., 111, 1996
|
|
2WU2
 
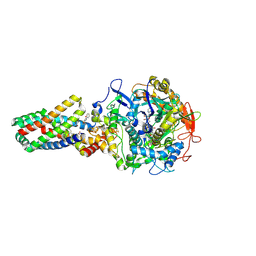 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhC His84Met mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-09-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E. Coli Succinate:Quinone Oxidoreductase (Sqr) Sdhc His84met Mutant
To be Published
|
|
3ET4
 
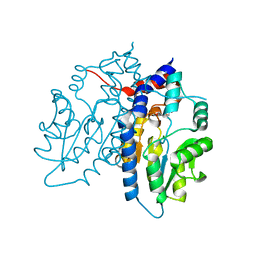 | | Structure of Recombinant Haemophilus Influenzae E(P4) Acid Phosphatase | | Descriptor: | MAGNESIUM ION, Outer membrane protein P4, NADP phosphatase, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Recombinant Haemophilus Influenzae E (P4) Acid Phosphatase
Reveals a New Member of the Haloacid Dehalogenase Superfamily.
Biochemistry, 46, 2007
|
|
3FK3
 
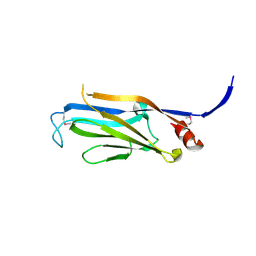 | | Structure of the Yeats Domain, Yaf9 | | Descriptor: | Protein AF-9 homolog | | Authors: | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | Deposit date: | 2008-12-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7N6S
 
 | |
7N56
 
 | |
4YNC
 
 | | OYE1 W116A COMPLEXED WITH (Z)-METHYL-3-CYANO-3-PHENYLACRYLATE IN A NON PRODUCTIVE BINDING MODE | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, ... | | Authors: | Santangelo, S, Brenna, E, Stewart, J.D, Powell III, R.W. | | Deposit date: | 2015-03-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Opposite Enantioselectivity in the Bioreduction of (Z)-beta-Aryl-beta-cyanoacrylates Mediated by the Tryptophan 116 Mutants of Old Yellow Enzyme 1: Synthetic Approach to (R)- and (S)-beta-Aryl-gamma-lactams
Adv.Synth.Catal., 357, 2015
|
|
8AHM
 
 | | Crystal structure of tubulin in complex with C(13)/C(13')-Bis-Desmethyl-Disorazole Z | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Oliva, M.A, Diaz, J.F, Altmann, K.H. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Synthesis and Biological Evaluation of C(13)/C(13')-Bis(desmethyl)disorazole Z.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2GUI
 
 | | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit of E. coli DNA Polymerase III | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA polymerase III epsilon subunit, ... | | Authors: | Park, A.Y, Carr, P.D, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2006-04-30 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit E. coli DNA Polymerase III
To be Published
|
|
4YIL
 
 | | OYE1 W116A COMPLEXED WITH (Z)-METHYL 3-CYANO-3-(4-FLUOROPHENYL)ACRYLATE IN A NON PRODUCTIVE BINDING MODE | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH dehydrogenase 1, ... | | Authors: | Santangelo, S, Brenna, E, Stewart, J.D, Powell III, R.W. | | Deposit date: | 2015-03-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Opposite Enantioselectivity in the Bioreduction of (Z)-beta-Aryl-beta-cyanoacrylates Mediated by the Tryptophan 116 Mutants of Old Yellow Enzyme 1: Synthetic Approach to (R)- and (S)-beta-Aryl-gamma-lactams
Adv.Synth.Catal., 357, 2015
|
|
5KTS
 
 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound citraconate and Fe4S4 cluster | | Descriptor: | (~{Z})-2-methylbut-2-enedioic acid, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
7MT4
 
 | | Crystal structure of tryptophan Synthase in complex with F9, NH4+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMMONIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MT5
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, CESIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1SIG
 
 | |
2G8Y
 
 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
7MT6
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, benzimidazole, pH7.8 - alpha aminoacrylate form - E(A-A)(BZI) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3TBI
 
 | |
3TX1
 
 | | X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
TO BE PUBLISHED
|
|
3C0U
 
 | | Crystal structure of E.coli yaeQ protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein yaeQ | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli yaeQ protein.
To be Published
|
|
3CUO
 
 | | Crystal structure of the predicted DNA-binding transcriptional regulator from E. coli | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygaV | | Authors: | Zhang, R, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the predicted DNA-binding transcriptional regulator from E. coli.
To be Published
|
|
3CYZ
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with 9-keto-2(E)-decenoic acid at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
1TMH
 
 | |
5QHR
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000635a | | Descriptor: | 1,2-ETHANEDIOL, N-(3-fluorophenyl)-5-methyl-1,3,4-thiadiazol-2-amine, Protein FAM83B | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5DWG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Triaryl-substituted Imine Analog, 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol | | Descriptor: | 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DXM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol | | Descriptor: | 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
