4MBB
 
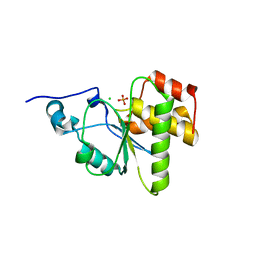 | | Cubic crystal form of PIR1 dual specificity phosphatase core | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Sankhala, R.S, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2013-08-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structure of Human PIR1, an Atypical Dual-Specificity Phosphatase.
Biochemistry, 53, 2014
|
|
3G5O
 
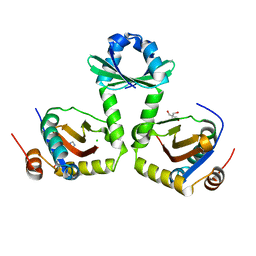 | | The crystal structure of the toxin-antitoxin complex RelBE2 (Rv2865-2866) from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
4IBU
 
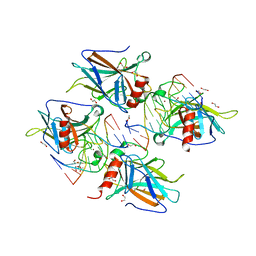 | | Human p53 core domain with hot spot mutation R273C and second-site suppressor mutation T284R in sequence-specific complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Eldar, A, Rozenberg, H, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
1ME3
 
 | |
4IDP
 
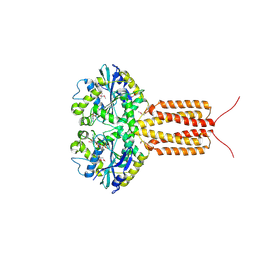 | | human atlastin-1 1-446, N440T, GppNHp | | Descriptor: | Atlastin-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Byrnes, L.J, Singh, A, Szeto, K, Benvin, N.M, O'Donnell, J.P, Zipfel, W.R, Sondermann, H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural basis for conformational switching and GTP loading of the large G protein atlastin.
Embo J., 32, 2013
|
|
1TU7
 
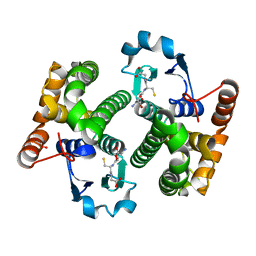 | |
3G8I
 
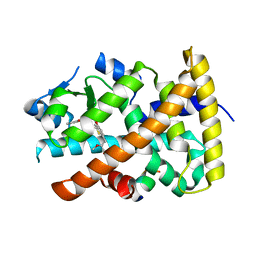 | | Aleglitazar, a new, potent, and balanced PPAR alpha/gamma agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puentener, K, Raab, S, Ruf, A, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3QBC
 
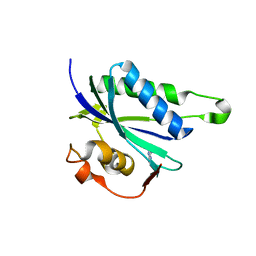 | |
7DLW
 
 | | Crystal structure of Arabidopsis ACS7 in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7, SULFATE ION | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
4DR9
 
 | | Crystal structure of a peptide deformylase from synechococcus elongatus in complex with actinonin | | Descriptor: | ACTINONIN, BROMIDE ION, Peptide deformylase, ... | | Authors: | Lorimer, D, Abendroth, J, Craig, T, Burgin, A, Segall, A, Rohwler, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
3QFH
 
 | | 2.05 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) from Staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, Epidermin leader peptide processing serine protease EpiP, GLYCEROL, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) from Staphylococcus aureus.
TO BE PUBLISHED
|
|
4ATT
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to a propynyl para methoxy phenyl | | Descriptor: | 3-(4-methoxyphenyl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
6A5T
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
1TWE
 
 | | INTERLEUKIN 1 BETA MUTANT F101Y | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Capsar, D.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
2XNB
 
 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 3,4-DIMETHYL-5-(2-{[(1Z)-4-PIPERAZIN-1-YLCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}PYRIMIDIN-4-YL)-1,3-THIAZOL-2(3H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-08-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
1C5C
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX | | Descriptor: | 2-ACETYLAMINO-NAPTHALENE-1,5-DISULFONIC ACID, CHIMERIC DECARBOXYLASE ANTIBODY 21D8, GLYCEROL | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-09 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
2BQW
 
 | | CRYSTAL STRUCTURE OF FACTOR XA IN COMPLEX WITH COMPOUND 45 | | Descriptor: | 1-{2-[(4-CHLOROPHENYL)AMINO]-2-OXOETHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-28 | | Release date: | 2006-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
1TWM
 
 | | Interleukin-1 Beta Mutant F146Y | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Capsar, D.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
2FYR
 
 | |
3KTL
 
 | | Crystal Structure of an I71A human GSTA1-1 mutant in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Achilonu, I.A, Gildenhuys, S, Fernandes, M.A, Fanucchi, S, Dirr, H.W. | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of a topologically conserved isoleucine in glutathione transferase structure, stability and function.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5PO9
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N07950b | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-phenyl-3-(trifluoromethyl)-1H-pyrazol-5-amine, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11039a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-{[(3R)-1-cyclopentyl-5-oxopyrrolidin-3-yl]methyl}methanesulfonamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1JER
 
 | | CUCUMBER STELLACYANIN, CU2+, PH 7.0 | | Descriptor: | COPPER (II) ION, CUCUMBER STELLACYANIN | | Authors: | Hart, P.J, Nersissian, A.M, Herrmann, R.G, Nalbandyan, R.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A missing link in cupredoxins: crystal structure of cucumber stellacyanin at 1.6 A resolution.
Protein Sci., 5, 1996
|
|
3KWF
 
 | | human DPP-IV with carmegliptin (S)-1-((2S,3S,11bS)-2-Amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl)-4-fluoromethyl-pyrrolidin-2-one | | Descriptor: | (4S)-1-[(2S,3S,11bS)-2-amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl]-4-(fluoromethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4AYR
 
 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
