7N5V
 
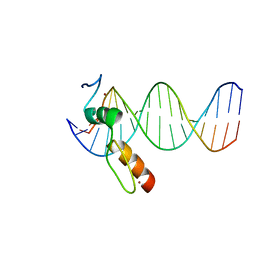 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7ORF
 
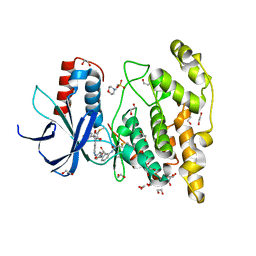 | | Crystal structure of JNK3 in complex with FMU-001-367 (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 10, ... | | Authors: | Chaikuad, A, Koch, P, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ORE
 
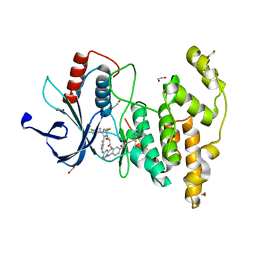 | | Crystal structure of JNK3 in complex with light-activated covalent inhibitor MR-II-249 with both non-covalent and covalent binding modes (compound 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[(5Z)-9-[[4-[5-(4-fluorophenyl)-3-methyl-2-methylsulfanyl-imidazol-4-yl]pyridin-2-yl]amino]-11,12-dihydrobenzo[c][1,2]benzodiazocin-2-yl]butanamide, Mitogen-activated protein kinase 10 | | Authors: | Chaikuad, A, Reynders, M, Trauner, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ORD
 
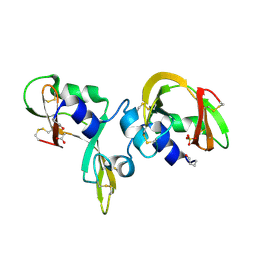 | |
7N5B
 
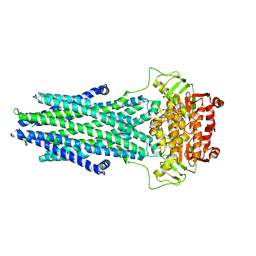 | | Structure of AtAtm3 in the outward-facing conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
7N58
 
 | | Structure of AtAtm3 in the inward-facing conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
7N59
 
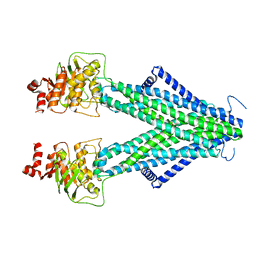 | | Structure of AtAtm3 in the inward-facing conformation with GSSG bound | | Descriptor: | ABC transporter B family member 25, mitochondrial, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
7N5A
 
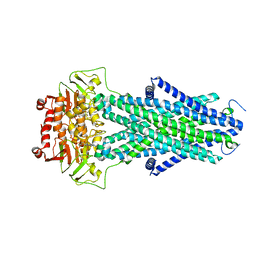 | | Structure of AtAtm3 in the closed conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
7OR6
 
 | |
7OR1
 
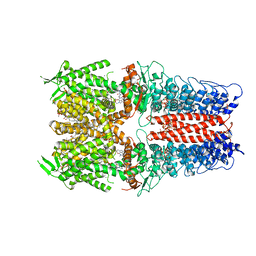 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
7OR0
 
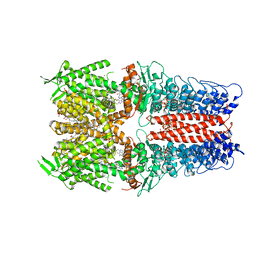 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
7N4N
 
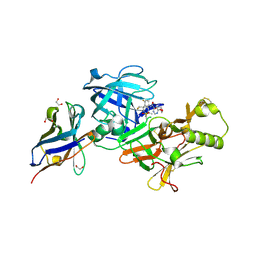 | | BACE-2 in complex with ligand 36 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 2, N-{3-[(2S,5R)-6-amino-2-(fluoromethyl)-5-(methanesulfonyl)-5-methyl-2,3,4,5-tetrahydropyridin-2-yl]-4-fluorophenyl}-6-methoxypyrimidine-4-carboxamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7N4Y
 
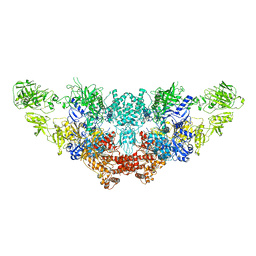 | | The structure of bovine thyroglobulin with iodinated tyrosines | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, K, Clarke, O.B. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | The structure of natively iodinated bovine thyroglobulin.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7N4T
 
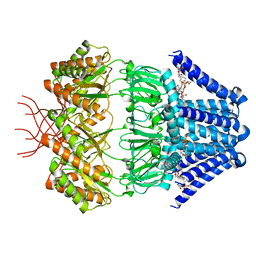 | | Low conductance mechanosensitive channel YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI | | Authors: | Catalano, C, Ben-Hail, D, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of Mechanosensitive Channel YnaI Using SMA2000: Challenges and Opportunities.
Membranes (Basel), 11, 2021
|
|
7N4P
 
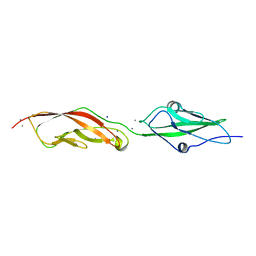 | | Crystal Structure of Lizard Cadherin-23 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin 23, SODIUM ION | | Authors: | Nisler, C.R, Sotomayor, M. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Interpreting the Evolutionary Echoes of a Protein Complex Essential for Inner-Ear Mechanosensation.
Mol.Biol.Evol., 40, 2023
|
|
7OQB
 
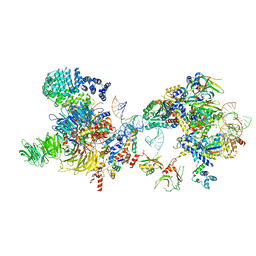 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQE
 
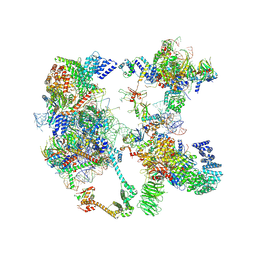 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQC
 
 | | The U1 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A), Pre-mRNA-processing factor 39, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQH
 
 | | CryoEM structure of the transcription termination factor Rho from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Saridakis, E, Vishwakarma, R, Lai Kee Him, J, Martin, K, Simon, I, Cohen-Gonsaud, M, Coste, F, Bron, P, Margeat, E, Boudvillain, M. | | Deposit date: | 2021-06-03 | | Release date: | 2022-02-09 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism.
Commun Biol, 5, 2022
|
|
7F09
 
 | | Crystal structure of the HLH-Lz domain of human TFE3 | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor E3, ZINC ION | | Authors: | Yang, G, Li, P, Liu, Z, Wu, S, Zhuang, C, Qiao, H, Fang, P, Wang, J. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the dimerization mechanism of human transcription factor E3.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
7F02
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7OPZ
 
 | | Camel GSTM1-1 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1.
Life, 12, 2022
|
|
7OPY
 
 | |
