4AAW
 
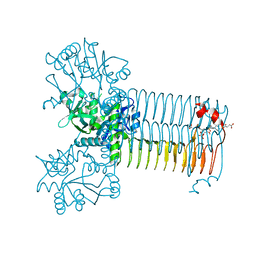 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | 4-{[1-(2-{[({5-[(3-carboxypropanoyl)amino]-2,4-dimethoxyphenyl}sulfonyl)amino]methyl}phenyl)piperidin-4-yl]methoxy}-4-oxobutanoic acid, BIFUNCTIONAL PROTEIN GLMU, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3R9A
 
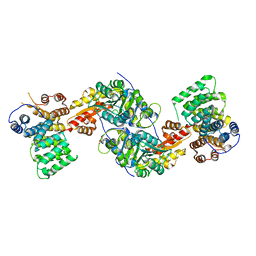 | | Human alanine-glyoxylate aminotransferase in complex with the TPR domain of human PEX5P | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peroxisomal targeting signal 1 receptor, Serine--pyruvate aminotransferase | | Authors: | Fodor, K, Wilmanns, M. | | Deposit date: | 2011-03-25 | | Release date: | 2011-05-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1
Plos Biol., 10, 2012
|
|
1L5V
 
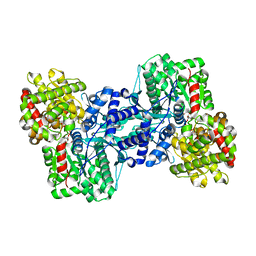 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with Glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MALTODEXTRIN PHOSPHORYLASE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
3FH1
 
 | |
3FFR
 
 | |
2ONF
 
 | |
3WUG
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
1REE
 
 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 3,4-EPOXYBUTYL-BETA-D-XYLOSIDE | | Descriptor: | (3S)-3-hydroxybutyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | Authors: | Rouvinen, J, Havukainen, R, Torronen, A. | | Deposit date: | 1995-12-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
2OX3
 
 | | R-state, PEP and Fru-6-P-bound, Escherichia coli fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, PHOSPHOENOLPYRUVATE | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of activated fructose-1,6-bisphosphatase from Escherichia coli. Coordinate regulation of bacterial metabolism and the conservation of the R-state.
J.Biol.Chem., 282, 2007
|
|
4HE0
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase | | Descriptor: | CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, MAGNESIUM ION, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3Q2U
 
 | |
4L6S
 
 | | PARP complexed with benzo[1,4]oxazin-3-one inhibitor | | Descriptor: | (2S)-6-{[4-(4-chlorophenyl)-3,6-dihydropyridin-1(2H)-yl]methyl}-2-methyl-2H-1,4-benzoxazin-3(4H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Dougan, D.R, Mol, C.D, Lawson, J.D. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel benzo[b][1,4]oxazin-3(4H)-ones as poly(ADP-ribose)polymerase inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3NDM
 
 | |
3B7V
 
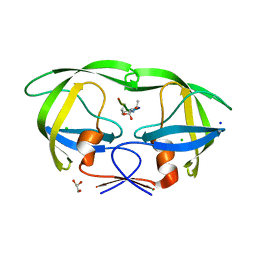 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate NLLTQI | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Kovalevsky, A.Y, Chumanevich, A.A, Weber, I.T. | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Caught in the Act: The 1.5 A Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate.
Biochemistry, 46, 2007
|
|
1H09
 
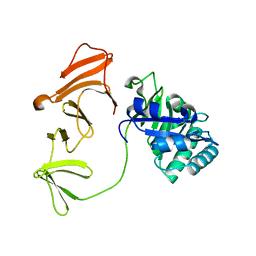 | | Multimodular Pneumococcal Cell Wall Endolysin from phage Cp-1 | | Descriptor: | LYSOZYME | | Authors: | Hermoso, J.A, Monterroso, B, Albert, A, Garcia, P, Menendez, M, Martinez-Ripoll, M, Garcia, J.L. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Selective Recognition of Pneumococcal Cell Wall by Modular Endolysin from Phage Cp-1
Structure, 11, 2003
|
|
6CDI
 
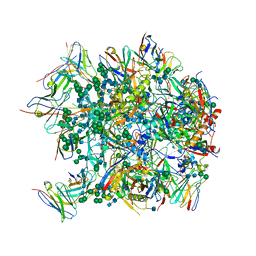 | | Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
1WJE
 
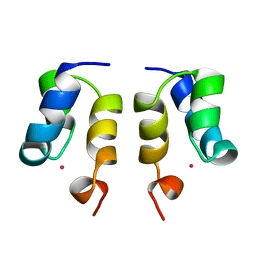 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1RLD
 
 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
3CU9
 
 | | High resolution crystal structure of 1,5-alpha-L-arabinanase from Geobacillus Stearothermophilus | | Descriptor: | CALCIUM ION, GLYCEROL, Intracellular arabinanase | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-04-16 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
4AC3
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-{2,4-DIMETHOXY-5-[(2-PIPERIDIN-1-YLBENZYL)sulfamoyl]phenyl}acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4JZZ
 
 | |
3PSU
 
 | |
2QL8
 
 | |
1ZEN
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Descriptor: | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE, ZINC ION | | Authors: | Cooper, S.J, Leonard, G.A, Hunter, W.N. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold.
Structure, 4, 1996
|
|
1HCU
 
 | | alpha-1,2-mannosidase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1,2-MANNOSIDASE, CALCIUM ION | | Authors: | Van Petegem, F, Contreras, H, Contreras, R, Van Beeumen, J. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Trichoderma Reesei Alpha-1,2-Mannosidase: Structural Basis for the Cleavage of Four Consecutive Mannose Residues
J.Mol.Biol., 312, 2001
|
|
