4R7A
 
 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
4O9D
 
 | | Structure of Dos1 propeller | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | Authors: | Kuscu, C, Schalch, T, Joshua-Tor, L. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2CRC
 
 | |
8A98
 
 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT | | Descriptor: | 40S ribosomal protein S12, 40S ribosomal protein S14, 40S ribosomal protein S19-like protein, ... | | Authors: | Rajan, K.S, Yonath, A, Bashan, A. | | Deposit date: | 2022-06-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8A3W
 
 | |
5H13
 
 | | EED in complex with PRC2 allosteric inhibitor EED396 | | Descriptor: | 4-azanylidene-2-(3-methoxy-4-propan-2-yloxy-phenyl)-6,7-dihydro-[1,3]benzodioxolo[6,5-a]quinolizine-3-carbonitrile, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
6FZC
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6G5I
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State R | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
5H17
 
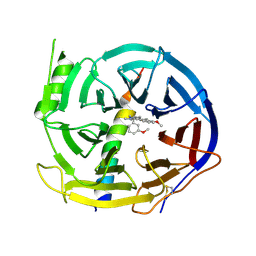 | | EED in complex with PRC2 allosteric inhibitor EED210 | | Descriptor: | (3R,4aS,10aS)-6-methoxy-3-[(3-methoxyphenyl)methyl]-1-methyl-3,4,4a,5,10,10a-hexahydro-2H-benzo[g]quinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
6G61
 
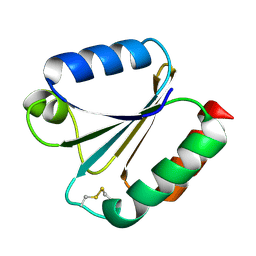 | |
6G6P
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.2 in P63 | | Descriptor: | Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
7O5B
 
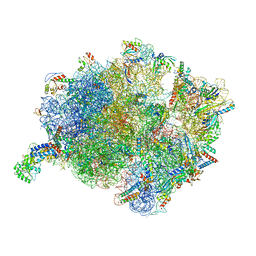 | | Cryo-EM structure of a Bacillus subtilis MifM-stalled ribosome-nascent chain complex with (p)ppGpp-SRP bound | | Descriptor: | 16S rRNA (1533-MER), 23S rRNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Czech, L, Berninghausen, O, Bange, G, Beckmann, R. | | Deposit date: | 2021-04-08 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
8AE6
 
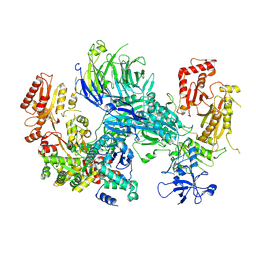 | | Cryo-EM structure of the SEA complex wing (SEACIT) | | Descriptor: | Maintenance of telomere capping protein 5, Nitrogen permease regulator 2, Nitrogen permease regulator 3, ... | | Authors: | Tafur, L, Loewith, R. | | Deposit date: | 2022-07-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the SEA complex.
Nature, 611, 2022
|
|
6FZ9
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2463 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
8AJM
 
 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJN
 
 | | Structure of the human DDB1-DCAF12 complex | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (30.6 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
6G18
 
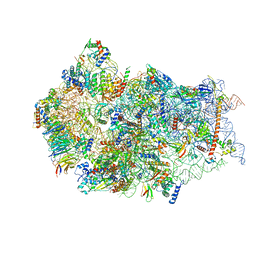 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State C | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
5HE2
 
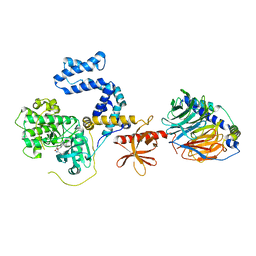 | | Bovine GRK2 in complex with Gbetagamma subunits and CCG224406 | | Descriptor: | (4~{S})-4-[3-[(2,6-dimethoxyphenyl)methylcarbamoyl]-4-fluoranyl-phenyl]-~{N}-(1~{H}-indazol-5-yl)-6-methyl-2-oxidanylidene-3,4-dihydro-1~{H}-pyrimidine-5-carboxamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cato, M.C, Waninger-Saroni, J, Tesmer, J.J.G. | | Deposit date: | 2016-01-05 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors.
J.Med.Chem., 59, 2016
|
|
6G5H
 
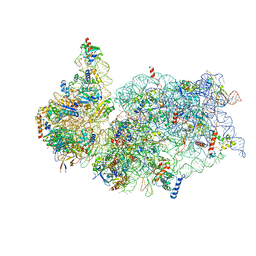 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - Mature | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G6M
 
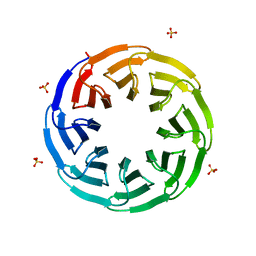 | | Crystal structure of the computationally designed Tako8 protein in P42212 | | Descriptor: | SULFATE ION, Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G6Q
 
 | | Crystal structure of the computationally designed Ika4 protein | | Descriptor: | Ika4 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6FYY
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6FZ7
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6G16
 
 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
