2EW5
 
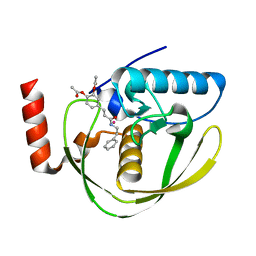 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | 4-{(1E)-3-OXO-3-[(2-PHENYLETHYL)AMINO]PROP-1-EN-1-YL}-1,2-PHENYLENE DIACETATE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2E3Q
 
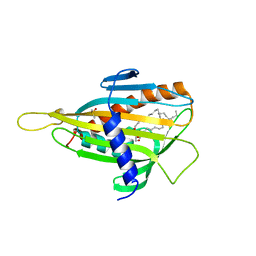 | | Crystal structure of CERT START domain in complex with C18-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3R
 
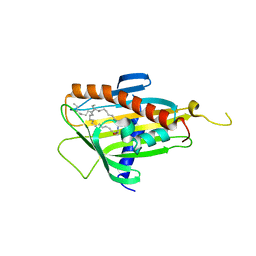 | | Crystal structure of CERT START domain in complex with C18-ceramide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3N
 
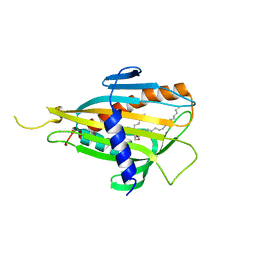 | | Crystal structure of CERT START domain in complex with C6-ceramide (P212121) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)HEXANAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3O
 
 | | Crystal structure of CERT START domain in complex with C16-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7G20
 
 | | Crystal Structure of human FABP4 in complex with 1-[(4-methoxyphenyl)methyl]-5-propan-2-yl-5-prop-2-enyl-1,3-diazinane-2,4,6-trione | | Descriptor: | (5S)-1-[(4-methoxyphenyl)methyl]-5-(propan-2-yl)-5-(prop-2-en-1-yl)-1,3-diazinane-2,4,6-trione, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Schnider, O, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FW2
 
 | | Crystal Structure of human FABP4 in complex with 5-[(4-chlorophenoxy)methyl]-4-prop-2-enyl-1,2,4-triazole-3-thiol | | Descriptor: | 5-[(4-chlorophenoxy)methyl]-4-(prop-2-en-1-yl)-4H-1,2,4-triazole-3-thiol, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8OQ8
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
7FZL
 
 | | Crystal Structure of human FABP4 in complex with 3-(3-chlorophenyl)-4-prop-2-enyl-1H-1,2,4-triazole-5-thione | | Descriptor: | (5P)-5-(3-chlorophenyl)-4-(prop-2-en-1-yl)-2,4-dihydro-3H-1,2,4-triazole-3-thione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8OQA
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
7FZV
 
 | | Crystal Structure of human FABP4 in complex with 1-[(2,4-dichlorophenyl)methyl]-4-hydroxy-3-[(E)-3-phenylprop-2-enyl]pyridin-2-one | | Descriptor: | 1-[(2,4-dichlorophenyl)methyl]-4-hydroxy-3-[(2E)-3-phenylprop-2-en-1-yl]pyridin-2(1H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8JYE
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JY9
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
7FYK
 
 | | Crystal Structure of human FABP4 in complex with 5-phenylmethoxy-4-prop-2-enyl-1,2,4-triazole-3-thiol | | Descriptor: | 5-(benzyloxy)-4-(prop-2-en-1-yl)-4H-1,2,4-triazole-3-thiol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8OKC
 
 | | SARS-CoV2 NSP5 in complex with a GC-376 based peptidomimetic PROTAC | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-1-[[(~{Z},2~{S})-5-[4-[[1-[2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]-6-fluoranyl-1,3-bis(oxidanylidene)isoindol-5-yl]piperidin-4-yl]methyl]piperazin-1-yl]-5-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]pent-3-en-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4CRT
 
 | | Crystal structure of human monoamine oxidase B in complex with the multi-target inhibitor ASS234 | | Descriptor: | (E)-N-methyl-N-[[1-methyl-5-[3-[1-(phenylmethyl)piperidin-4-yl]propoxy]indol-2-yl]methyl]prop-1-en-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Esteban, G, Allan, J, Samadi, A, Mattevi, A, Unzeta, M, Marco-Contelles, J, Binda, C, Ramsay, R.R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-04-02 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Analysis of the Irreversible Inhibition of Human Monoamine Oxidases by Ass234, a Multi-Target Compound Designed for Use in Alzheimer'S Disease.
Biochim.Biophys.Acta, 1844, 2014
|
|
7H71
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1416571195 (CHIKV_MacB-x0441) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(cyclopent-3-en-1-yl)amino]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
8OI3
 
 | | Structure of NopD with AtSUMO2 | | Descriptor: | Small ubiquitin-related modifier 2, Type III effector, prop-2-en-1-amine | | Authors: | Reverter, D, Li, Y. | | Deposit date: | 2023-03-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of NopD with AtSUMO2
To Be Published
|
|
7K65
 
 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JXT
 
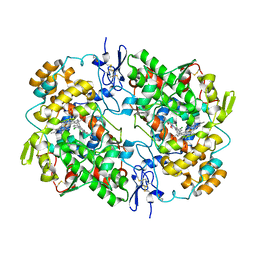 | | Ovine COX-1 in complex with the subtype-selective derivative 2a | | Descriptor: | 2-[4,5-bis(2-chlorophenyl)-1H-imidazol-2-yl]-6-(prop-2-en-1-yl)phenyl methoxyacetate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ko, Y, Iaselli, M, Miciaccia, M, Friedrich, L, Schneider, G, Scilimati, A, Cingolani, G. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Learning from Nature: From a Marine Natural Product to Synthetic Cyclooxygenase-1 Inhibitors by Automated De Novo Design.
Adv Sci, 8, 2021
|
|
7K9T
 
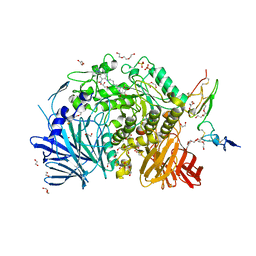 | | Co-crystal structure of alpha glucosidase with compound 5 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[(5Z)-6-{[2-nitro-4-(2H-1,2,3-triazol-2-yl)phenyl]amino}hex-5-en-1-yl]amino}cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
4D9T
 
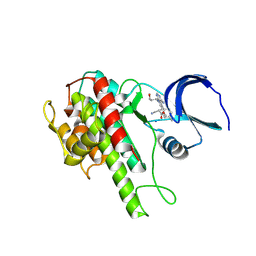 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
