7YXV
 
 | |
6XF4
 
 | | Crystal structure of STING REF variant in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), 1,2-ETHANEDIOL, Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6XF3
 
 | | Crystal structure of STING in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
7AG0
 
 | | Complex between the bone morphogenetic protein 2 and its antagonist Noggin | | Descriptor: | Bone morphogenetic protein 2, GLYCEROL, Noggin | | Authors: | Robert, C, Bruck, F, Herman, R, Vandevenne, M, Filee, P, Kerff, F, Matagne, A. | | Deposit date: | 2020-09-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural analysis of the interaction between human cytokine BMP-2 and the antagonist Noggin reveals molecular details of cell chondrogenesis inhibition.
J.Biol.Chem., 299, 2023
|
|
1CA1
 
 | |
2RJI
 
 | |
6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6S26
 
 | | Crystal structure of human wild type STING in complex with 2'-3'-cyclic-GMP-7-deaza-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M, Brynda, J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic Preparation of 2'-5',3'-5'-Cyclic Dinucleotides, Their Binding Properties to Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations.
J.Med.Chem., 62, 2019
|
|
8U00
 
 | | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides
To Be Published
|
|
3VKW
 
 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
6S86
 
 | | Human wtSTING in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of Interferon genes | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2019-07-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | No magnesium is needed for binding of the stimulator of interferon genes to cyclic dinucleotides.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
152D
 
 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
5FRA
 
 | | CBM40_CPF0721-6'SL | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Ribeiro, J.P, Pau, W, Pifferi, C, Renaudet, O, Varrot, A, Mahal, L.K, Imberty, A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a High-Affinity Sialic Acid-Specific Cbm40 from Clostridium Perfringens and Engineering of a Divalent Form.
Biochem.J., 473, 2016
|
|
3L22
 
 | |
2X79
 
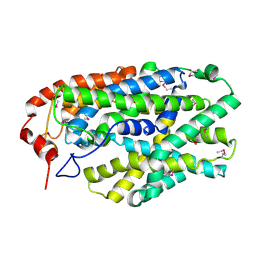 | | Inward facing conformation of Mhp1 | | Descriptor: | HYDANTOIN TRANSPORT PROTEIN | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
6S27
 
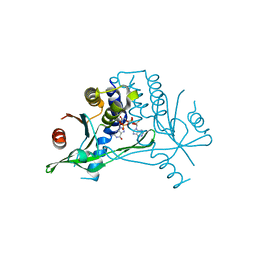 | |
7BXY
 
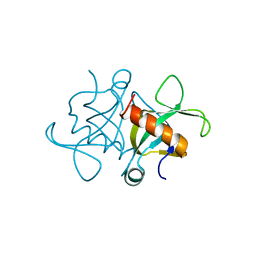 | | mRNA interferase from Bacillus cereus | | Descriptor: | mRNA interferase | | Authors: | Kang, S.M. | | Deposit date: | 2020-04-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | mRNA interferase from Bacillus cereus
To Be Published
|
|
151D
 
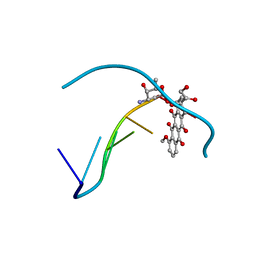 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
2RG2
 
 | |
2RLC
 
 | |
5I8F
 
 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
3IJM
 
 | | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale. | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Cuff, M.E, Tesar, C, Samano, S, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale.
TO BE PUBLISHED
|
|
3IUZ
 
 | |
3JQ1
 
 | |
3JQ0
 
 | |
