3FGV
 
 | |
3G0J
 
 | | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1)
To be Published
|
|
2ZIC
 
 | | Crystal structure of Streptococcus mutans dextran glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Dextran glucosidase, ... | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
2SLI
 
 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC, THE REACTION PRODUCT | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
1Z9T
 
 | |
2RF5
 
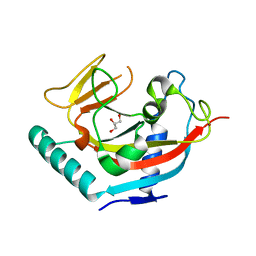 | | Crystal structure of human tankyrase 1- catalytic PARP domain | | Descriptor: | GLYCEROL, Tankyrase-1, ZINC ION | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Zinc binding catalytic domain of human tankyrase 1.
J.Mol.Biol., 379, 2008
|
|
3GF6
 
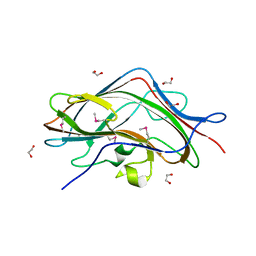 | |
1XLC
 
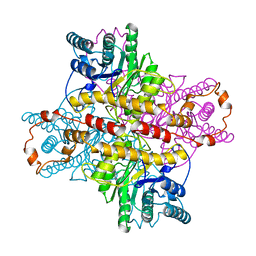 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
3OSM
 
 | |
3OSH
 
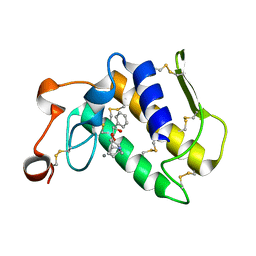 | | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Shukla, P.K, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-09 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution
To be Published
|
|
6DFF
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef monomer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
3DFE
 
 | |
1XYP
 
 | |
1XLF
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, D-gluconic acid, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
3AKI
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido | | Descriptor: | (2R,3R,4R,5S)-2-azido-5-(hydroxymethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3OT2
 
 | |
3DFO
 
 | |
1WJA
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
3GGD
 
 | |
4I2P
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with rilpivirine (TMC278) based analogue | | Descriptor: | (2E)-3-[4-({6-[(4-methoxyphenyl)amino]-7H-purin-2-yl}amino)-3,5-dimethylphenyl]prop-2-enenitrile, Gag-Pol polyprotein | | Authors: | Patel, D, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2012-11-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2964 Å) | | Cite: | A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9, 2012
|
|
3G3S
 
 | |
3GD0
 
 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase) | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
4GYQ
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase NDM-1, MAGNESIUM ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
