5K8D
 
 | | Crystal structure of rFVIIIFc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Leksa, N, Quan, C. | | Deposit date: | 2016-05-29 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | The structural basis for the functional comparability of factor VIII and the long-acting variant recombinant factor VIII Fc fusion protein.
J. Thromb. Haemost., 15, 2017
|
|
3L8Q
 
 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
3UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, CMP, AND ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
8XT1
 
 | | Cryo-EM structure of the human 39S mitoribosome with 5uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT3
 
 | | Cryo-EM structure of the human 39S mitoribosome with 10uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT2
 
 | | Cryo-EM structure of the human 55S mitoribosome with 10uM Tigecycline | | Descriptor: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT0
 
 | | Cryo-EM structure of the human 55S mitoribosome with 5um Tigecycline | | Descriptor: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
3LAD
 
 | |
8VJ7
 
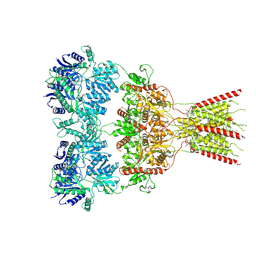 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, GLUTAMIC ACID, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.85 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
9BER
 
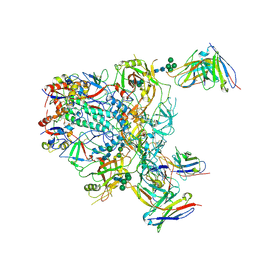 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
6VP6
 
 | |
6WLN
 
 | | hc16 ligase product models, 10.0 Angstrom resolution | | Descriptor: | RNA (349-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
3PPS
 
 | | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria--common structural features of asco-laccases.
Febs J., 278, 2011
|
|
9B17
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PAN1 inhibitor | | Descriptor: | (5P)-5-(1H-indol-3-yl)-1-[2-(piperazin-1-yl)ethyl]-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
9B1Q
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PYN1 inhibitor | | Descriptor: | (5P)-1-[(imidazolidin-1-yl)methyl]-5-(1H-indol-3-yl)-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
4L1E
 
 | | Crystal structure of C-Phycocyanin from Leptolyngbya sp. N62DM | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, Phycocyanin alpha chain, ... | | Authors: | Singh, N.K, Raj, I, Gourinath, S, Madamwar, D. | | Deposit date: | 2013-06-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure and Interaction of Phycocyanin with beta-Secretase: A Putative Therapy for Alzheimer's Disease.
CNS Neurol Disord Drug Targets, 13, 2014
|
|
721P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8B3X
 
 | | High resolution crystal structure of dimeric SUDV VP40 | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Norris, M, Saphire, E.O, Becker, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
3PE0
 
 | |
8FCS
 
 | |
1EJD
 
 | | Crystal structure of unliganded mura (type1) | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1EXW
 
 | |
1EA7
 
 | | Sphericase | | Descriptor: | CALCIUM ION, SERINE PROTEASE | | Authors: | Almog, O, Gonzalez, A, Klein, D, Braun, S, Shoham, G. | | Deposit date: | 2001-07-10 | | Release date: | 2002-07-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The 0.93A Crystal Structure of Sphericase: A Calcium-Loaded Serine Protease from Bacillus Sphaericus
J.Mol.Biol., 332, 2003
|
|
1E5A
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | 2,4,6-TRIBROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
