2KWZ
 
 | | Solution structure of NS2 [60-99] | | Descriptor: | Protease NS2-3 | | Authors: | Montserret, R, Bartenschlager, R, Penin, F. | | Deposit date: | 2010-04-22 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly.
Plos Pathog., 6, 2010
|
|
3L3L
 
 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2LZP
 
 | |
1W8C
 
 | |
8OK2
 
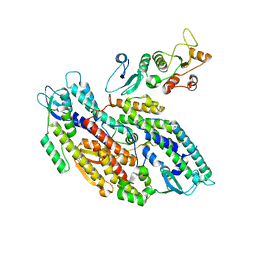 | | Bipartite interaction of TOPBP1 with the GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, DNA replication complex GINS protein PSF3, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2023-03-26 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | TopBP1 utilises a bipartite GINS binding mode to support genome replication.
Nat Commun, 15, 2024
|
|
2MKB
 
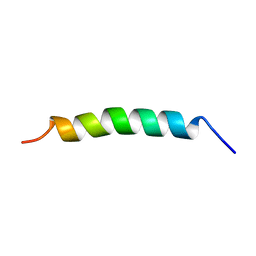 | |
2LZQ
 
 | |
1NG9
 
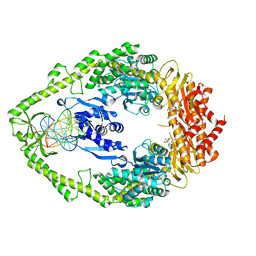 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2002-12-17 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
4L6S
 
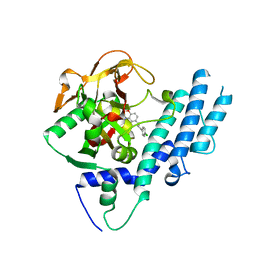 | | PARP complexed with benzo[1,4]oxazin-3-one inhibitor | | Descriptor: | (2S)-6-{[4-(4-chlorophenyl)-3,6-dihydropyridin-1(2H)-yl]methyl}-2-methyl-2H-1,4-benzoxazin-3(4H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Dougan, D.R, Mol, C.D, Lawson, J.D. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel benzo[b][1,4]oxazin-3(4H)-ones as poly(ADP-ribose)polymerase inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2LPB
 
 | | Structure of the complex of the central activation domain of Gcn4 bound to the mediator co-activator domain 1 of Gal11/med15 | | Descriptor: | General control protein GCN4, Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Brzovic, P.S, Heikaus, C.C, Kisselev, L, Vernon, R, Herbig, E, Pacheco, D, Warfield, L, Littlefield, P, Baker, D, Klevit, R.E, Hahn, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Mol.Cell, 44, 2011
|
|
2K0N
 
 | |
4CFM
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 6-(cyclohexylmethoxy)-8-(2-methylphenyl)-9H-purin-2-amine, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFW
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylbenzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFU
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-azanyl-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methyl-benzoic acid, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFV
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylphenol, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFN
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(cyclohexylmethoxy)-8-(trifluoromethyl)-9H-purin-2-amine, CYCLIN-A2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFX
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]benzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
3K77
 
 | | X-ray crystal structure of XRCC1 | | Descriptor: | DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, London, R.E. | | Deposit date: | 2009-10-12 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LQK
 
 | |
3HH2
 
 | |
3H15
 
 | |
1HKQ
 
 | | PPS10 plasmid DNA replication initiator protein RepA. Replication inactive, dimeric N-terminal domain. | | Descriptor: | BENZOIC ACID, MERCURY (II) ION, PHOSPHATE ION, ... | | Authors: | Giraldo, R, Fernandez-Tornero, C, Evans, P.R, Diaz-Orejas, R, Romero, A. | | Deposit date: | 2003-03-11 | | Release date: | 2003-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Conformational Switch between Transcriptional Repression and Replication Initiation in Repa Dimerization Domain
Nat.Struct.Biol., 10, 2003
|
|
3GPN
 
 | | Structure of the non-trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GPM
 
 | | Structure of the trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2WVR
 
 | | Human Cdt1:Geminin complex | | Descriptor: | DNA REPLICATION FACTOR CDT1, GEMININ | | Authors: | De Marco, V, Perrakis, A. | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Quaternary Structure of the Human Cdt1-Geminin Complex Regulates DNA Replication Licensing.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
