4BPU
 
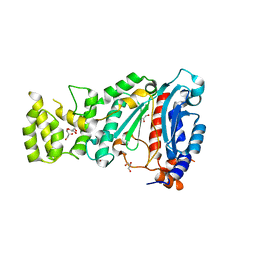 | | Crystal structure of human primase in heterodimeric form, comprising PriS and truncated PriL lacking the C-terminal Fe-S domain. | | Descriptor: | DNA PRIMASE LARGE SUBUNIT, DNA PRIMASE SMALL SUBUNIT, GLYCEROL, ... | | Authors: | Kilkenny, M.L, Perera, R.L, Pellegrini, L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Human Primase Reveal Design of Nucleotide Elongation Site and Mode of Pol Alpha Tethering
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BY2
 
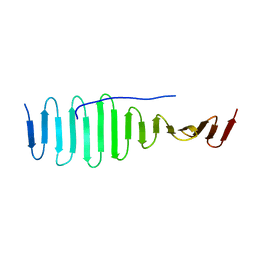 | | SAS-4 (dCPAP) TCP domain in complex with a Proline Rich Motif of Ana2 (dSTIL) of Drosophila Melanogaster | | Descriptor: | 1,2-ETHANEDIOL, ANASTRAL SPINDLE 2, SAS 4 | | Authors: | Cottee, M.A, Muschalik, N, Wong, Y.L, Johnson, C.M, Johnson, S, Andreeva, A, Oegema, K, Lea, S.M, Raff, J.W, van Breugel, M. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly.
Elife, 2, 2013
|
|
4C95
 
 | |
4CBP
 
 | | Crystal structure of neural ectodermal development factor IMP-L2. | | Descriptor: | GLYCEROL, NEURAL/ECTODERMAL DEVELOPMENT FACTOR IMP-L2 | | Authors: | Kulahin, N, Kristensen, O, Brzozowski, M, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-10-29 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis of Imp-L2 Function
To be Published
|
|
4C33
 
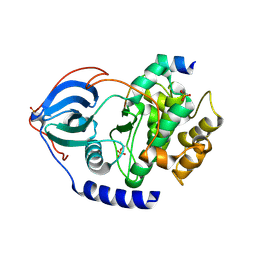 | | PKA-S6K1 Chimera Apo | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
4BXW
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | COAGULATION FACTOR V, FACTOR XA, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-16 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
4C4I
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(dimethylcarbamoyl)phenyl]amino}-2-(1,3-oxazol-5-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C9J
 
 | | Structure of yeast mitochondrial ADP/ATP carrier isoform 3 inhibited by carboxyatractyloside (P212121 crystal form) | | Descriptor: | ADP, ATP CARRIER PROTEIN 3, CARDIOLIPIN, ... | | Authors: | Ruprecht, J.J, Hellawell, A.M, Harding, M, Crichton, P.G, McCoy, A.J, Kunji, E.R.S. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.397 Å) | | Cite: | Structures of Yeast Mitochondrial Adp/ATP Carriers Support a Domain-Based Alternating-Access Transport Mechanism
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
4CBC
 
 | |
4C9H
 
 | | Structure of yeast mitochondrial ADP/ATP carrier isoform 2 inhibited by carboxyatractyloside (P212121 crystal form) | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, ADP, ATP CARRIER PROTEIN 2, ... | | Authors: | Ruprecht, J.J, Hellawell, A.M, Harding, M, Crichton, P.G, McCoy, A.J, Kunji, E.R.S. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Yeast Mitochondrial Adp/ATP Carriers Support a Domain-Based Alternating-Access Transport Mechanism
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4C08
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with CaCl2 at 1.34 Angstroms | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.338 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
4C4J
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(1-methyl-1H-imidazol-5-yl)phenyl]amino}-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C8D
 
 | | Crystal structure of JmjC domain of human histone 3 Lysine-specific demethylase 3B (KDM3B) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vollmar, M, Johansson, C, Gileadi, C, Goubin, S, Szykowska, A, Krojer, T, Crawley, L, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone 3 Lysine-Specific Demethylase 3B (Kdm3B)
To be Published
|
|
4BW5
 
 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CADMIUM ION, POTASSIUM CHANNEL SUBFAMILY K MEMBER 10, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Dong, L, Quigley, A, Shrestha, L, Mukhopadhyay, S, Strain-Damerell, C, Goubin, S, Grieben, M, Shintre, C.A, Mackenzie, A, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2013-06-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | K2P Channel Gating Mechanisms Revealed by Structures of Trek-2 and a Complex with Prozac
Science, 347, 2015
|
|
4C06
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with MgCl2 | | Descriptor: | MAGNESIUM ION, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
4C3Z
 
 | | Nucleotide-free crystal structure of nucleotide-binding domain 1 from human MRP1 supports a general-base catalysis mechanism for ATP hydrolysis. | | Descriptor: | MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 1, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Magnard, S, Falson, P, Di Pietro, A, Baubichon-Cortay, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleotide-Free Crystal Structure of Nucleotide-Binding Domain 1 from Human Abcc1 Supports a 'General-Base Catalysis' Mechanism for ATP Hydrolysis.
Biochem.Pharm., 3, 2014
|
|
4C05
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with SAH | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
4C1I
 
 | | Selective Inhibitors of PDE2, PDE9, and PDE10: Modulators of Activity of the Central Nervous System | | Descriptor: | (2S,3R)-3-(6-amino-9H-purin-9-yl)nonan-2-ol, CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Jorgensen, M, Kehler, J, Langgard, M, Svenstrup, N, Tagmose, L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chapter 4: Selective Inhibitors of Pde2, Pde9, and Pde10: Modulators of Activity of the Central Nervous System
To be Published
|
|
4C4W
 
 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | Descriptor: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
4CBA
 
 | | Structural of delta 1-76 CTNNBL1 in space group I222 | | Descriptor: | 1,2-ETHANEDIOL, BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha.
Febs Lett., 588, 2014
|
|
4C6R
 
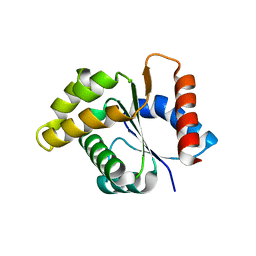 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPS4 | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
4C8B
 
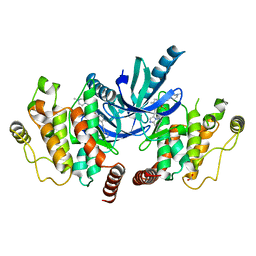 | | Structure of the kinase domain of human RIPK2 in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Canning, P, Krojer, T, Bradley, A, Mahajan, P, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Inflammatory Signaling by NOD-RIPK2 Is Inhibited by Clinically Relevant Type II Kinase Inhibitors.
Chem. Biol., 22, 2015
|
|
5OWC
 
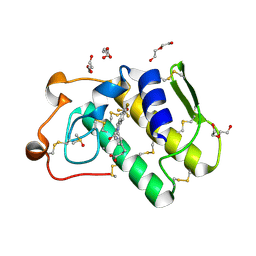 | | Indole-2 carboxamides as selective secreted phospholipase A2 type X (sPLA2-X) inhibitors | | Descriptor: | 3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]propanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J.S, Roth, R.G, Knerr, L, Bodin, C, Pettersen, D. | | Deposit date: | 2017-08-31 | | Release date: | 2018-08-01 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Series of Indole-2 Carboxamides as Selective Secreted Phospholipase A2Type X (sPLA2-X) Inhibitors.
Acs Med.Chem.Lett., 9, 2018
|
|
4CB9
 
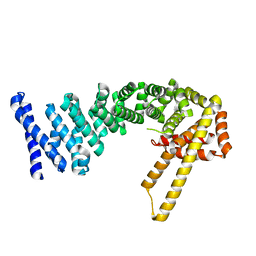 | | Structure of full-length CTNNBL1 in P43212 space group | | Descriptor: | BETA-CATENIN-LIKE PROTEIN 1 | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mutational Analysis Reveals that Ctnnbl1 Binds Nlss in a Manner Distinct from that of its Closest Armadillo-Relative, Karyopherin Alpha
FEBS Lett., 588, 2014
|
|
