5TRK
 
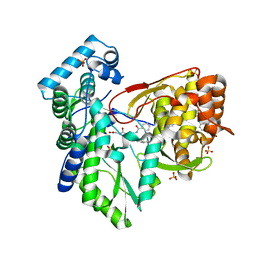 | |
6MEP
 
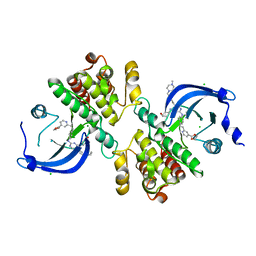 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
3RJT
 
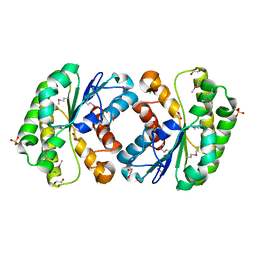 | | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
8P1G
 
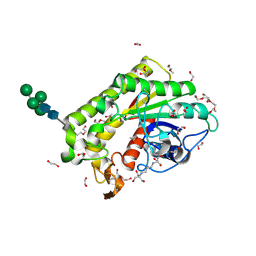 | | Crystal structure of inactive TtCE16 in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Kosinas, C, Pentari, C, Zerva, A, Topakas, E, Karampa, P. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The role of CE16 exo-deacetylases in hemicellulolytic enzyme mixtures revealed by the biochemical and structural study of the novel TtCE16B esterase.
Carbohydr Polym, 327, 2024
|
|
4D2T
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-[2-(phenylcarbamoyl)-5-(1H-pyrazol-4-yl)phenoxy]propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-13 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
3LU6
 
 | | Human serum albumin in complex with compound 1 | | Descriptor: | Serum albumin, [(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]acetic acid | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
3RNJ
 
 | | Crystal structure of the SH3 domain from IRSp53 (BAIAP2) | | Descriptor: | 1,2-ETHANEDIOL, Brain-specific angiogenesis inhibitor 1-associated protein 2, ISOPROPYL ALCOHOL, ... | | Authors: | Simister, P.C, Barilari, M, Muniz, J.R.C, Dente, L, Knapp, S, von Delft, F, Filippakopoulos, P, Vollmar, M, Chaikuad, A, Raynor, J, Tregubova, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the SH3 domain from IRSp53 (BAIAP2)
To be Published
|
|
4ZEB
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
6LSR
 
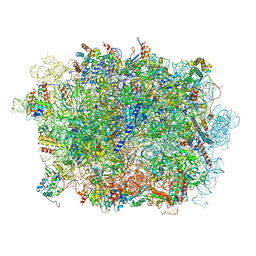 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
4L42
 
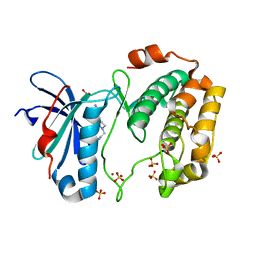 | | Crystal structures of human p70S6K1-PIF | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, SULFATE ION, ... | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4L4W
 
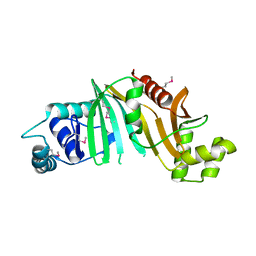 | |
7B6S
 
 | | Sheep Polyomavirus VP1 in complex with 10 mM Forssman antigen pentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
2A2G
 
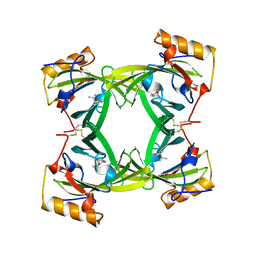 | | THE CRYSTAL STRUCTURES OF A2U-GLOBULIN AND ITS COMPLEX WITH A HYALINE DROPLET INDUCER. | | Descriptor: | D-LIMONENE 1,2-EPOXIDE, PROTEIN (ALPHA-2U-GLOBULIN) | | Authors: | Chaudhuri, B.N, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1998-11-19 | | Release date: | 1999-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structures of alpha 2u-globulin and its complex with a hyaline droplet inducer.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3G9E
 
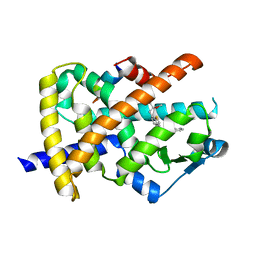 | | Aleglitaar. a new. potent, and balanced dual ppara/g agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Ruf, A, Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puenterner, K, Raab, S, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6HQF
 
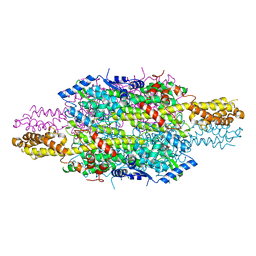 | | Structure of Phenylalanine ammonia-lyase from Petroselinum crispum in complex with (R)-APEP | | Descriptor: | Phenylalanine ammonia-lyase 1, [(1R)-1-amino-2-phenylethyl]phosphonic acid | | Authors: | Bata, Z, Molnar, B, Leveles, I, Poppe, L, Vertessy, G.B. | | Deposit date: | 2018-09-24 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Substrate Tunnel Engineering Aided by X-ray Crystallography and Functional Dynamics Swaps the Function of MIO-Enzymes
Acs Catalysis, 2021
|
|
4OGB
 
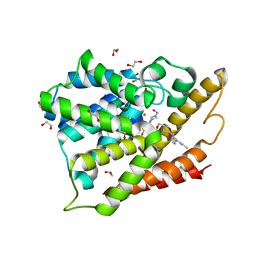 | | Crystal structure of the catalytic domain of PDE4D2 with compound 2 | | Descriptor: | (2R)-8-(3,4-dimethoxyphenyl)-6-methyl-2-(tetrahydro-2H-pyran-4-yl)-2H-chromen-4-ol, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feil, S.C, Parker, M.W. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | The PDE inhibition profile of LY294002 and tetrahydropyranyl analogues reveals a chromone motif for the development of PDE inhibitors
To be Published
|
|
1GDQ
 
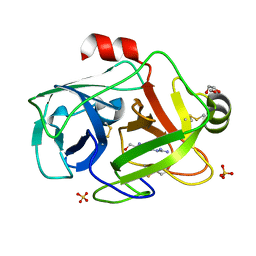 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1LL7
 
 | |
6C63
 
 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.900028 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
1LP4
 
 | | Crystal structure of a binary complex of the catalytic subunit of protein kinase CK2 with Mg-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-07 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
7JV8
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with compound 35 | | Descriptor: | 5'-nucleotidase, 6-chloro-N-cyclopentyl-1-{5-O-[(2R)-1-hydroxy-3-methoxy-2-phosphonopropan-2-yl]-beta-D-ribofuranosyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Gibbons, P, Du, X. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Orally Bioavailable Small-Molecule CD73 Inhibitor (OP-5244) Reverses Immunosuppression through Blockade of Adenosine Production.
J.Med.Chem., 63, 2020
|
|
4H3F
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
1VQ0
 
 | |
